An introduction to the genetic code
The sequence of bases in the mRNA molecule specifies the order of the amino acids to be assembled by the ribosome. But it’s not a 1 to 1 translation of base to amino acid. There are only four nucleotides, but there are 20 amino acids to encode. So early on, molecular biologists hypothesized that the genetic code must read multiple bases together as a unit, called a codon.
The code is degenerate.
The genetic code uses three-base codons, reading 5’ to 3’ along the RNA. There are 64 possible combinations of three bases and 64 codons. 61 of these codons specify amino acids, and 3 are used as stop codons to mark the end of the coding sequence of a protein. Because there are 64 codons but only 20 amino acids, there are extra codons to go around and some amino acids are specified by more than one codon. Because of this, the genetic code is said to be degenerate.
The code is universal.
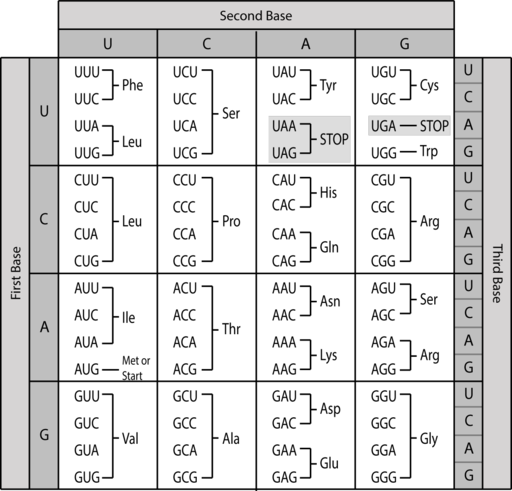
The code is also universal – with only a few very rare exceptions, the meaning of these codons is consistent among all living organisms. A codon table is shown in Figure 8. All 64 possible combinations of bases are listed, organized by the first base of the codon in four rows, the second base of the codon in four columns, and the third base of the codon on different lines in each of the 16 boxes of the table. The codons are read 5’ to 3’, left to right, so a GUU valine codon could be more completely described as 5’GUU3’.
There are four special codons to notice: First, the codon AUG is labeled “Met or Start”. AUG specifies the amino acid methionine. AUG is also the first codon to be translated in nearly every protein, which is why it is called the start codon. Methionine is thus the first amino acid incorporated into nearly every protein, although in prokaryotes a specially modified methionine called formyl-methionine (or f-Met) is used as the initiating amino acid. AUG codons can also be found in the middle of a coding sequence, where they also specify methionine.
Second, the codons UAA, UAG, and UGA are labeled “stop”. These codons do not specify an amino acid. Instead, they are used to signal the end of a protein-coding sequence.
Test Your Understanding
The code is non-overlapping
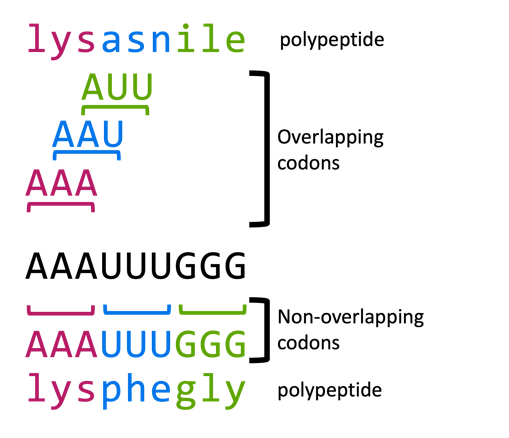
The genetic code is read three bases at a time, with each codon immediately adjacent to the next. There are no “spacer” bases, and codons do not overlap. This is illustrated in Figure 9. The protein is synthesized from N- to C- terminus. So the RNA sequence AAAUUUGGG might be read in non-overlapping codons of AAA-UUU-GGG, encoding the protein N-lys-phe-glu-C.
Why “might” be read? Because the codons are non-overlapping, it also means that for every RNA, there are three potential reading frames, depending on which base you start reading with. For the sequence of AAAUUUGGG shown in Figure 9, the three potential reading frames would be:
AAA-UUU-GGG (lys-phe-gly)
A-AAU-UUG-GG (-asn-leu)
AA-AUU-UGG-G (-lle-trp)
These reading frames encode very different polypeptides!
Note that an mRNA is not read beginning with the first amino acid. There is always an untranslated region (UTR) at the 5’ end of the molecule. It is the start codon that establishes the reading frame for an RNA. If you do not know the reading frame for an RNA, then you cannot determine the sequence of the protein, even if you know the RNA sequence.
The amino acid-specifying codons are read in frame to specify the sequence of amino acids. The coding sequence ends with a stop codon, but the RNA continues past that point. Just as every RNA molecule has a 5’ UTR, every RNA also has a 3’UTR.
Test Your Understanding
Media Attributions
- Genetic Code © Wikipedia is licensed under a CC0 (Creative Commons Zero) license
- Non-overlapping © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license

