Chromatin and Chromatin Dynamics
Typically, we think of our genome as static, with genetic information passed along mostly intact from generation to generation. However, the overall structure of chromatin is not static. It changes by the stage of the cell cycle, with chromosomes condensing in preparation for mitosis or meiosis. It also changes in response to other stimuli affecting gene regulation. Different parts of the genome are packaged differently within chromatin. The Genome Structure module described how areas of euchromatin and heterochromatin – loosely packed and tightly condensed, respectively – can be observed microscopically.
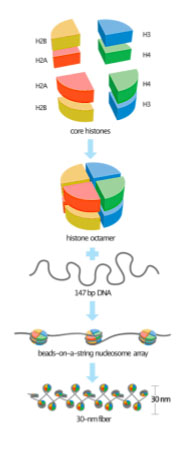
Recall from the module on Genome Structure that chromatin consists of DNA wrapped around packaging proteins, which allow meters of DNA to be condensed into a nucleus that is measured in micrometers. Even the least condensed DNA in the cell is wrapped around histone proteins, forming a nucleosome core, as shown in Figure 2. The nucleosome core consists of about 147 base pairs of DNA wrapped around eight histone proteins: two copies each of histones H2A, H2B, H3, and H4. Because of this, the core histones are called a histone octamer. As illustrated in the bottom two panels, the least-condensed chromatin has the appearance of beads on a string when viewed using an electron microscope, but a chromosome is further condensed by packing nucleosomes tightly together. Not shown in this image is the linker histone H1, which helps pack nucleosomes more tightly, and other non-histone proteins that affect higher-order packing.
Test Your Understanding
Three views of the molecular structure of a nucleosome are shown in Figure 3. The left panel shows a space-filling model of the nucleosome, with DNA shown in grey. The middle panel shows a ribbon diagram of the proteins, with DNA shown in purple. The right panel shows the same structure as in the middle but rotated 90 degrees to view the barrel-shaped structure from “above.” In all three panels, histone H2A is shown in red, H2B in yellow, H3 in blue, and H4 in green.
Also recall: The modules on transcription and gene expression discussed how eukaryotic genes are regulated via promoter elements and enhancers. Protein factors bind to those DNA elements to regulate expression. But the proteins of chromatin can interfere with or block that binding. Usually, only short stretches of unbound DNA are accessible to proteins involved in cell functions like transcription. The rest wraps around histones, and those histones must be circumvented for the cell to access the DNA for gene regulation. The DNA is unwrapped from the histones, or the histones are moved before the DNA double helix is melted for replication. A class of proteins called chromatin remodelers helps make this happen.
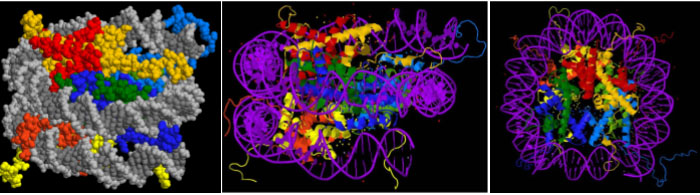
In the nucleosome structure shown in Figure 3, you can see that the bulk of each protein fits in the center of the coiled DNA. But the ends of the histone polypeptides – called the histone tails – stick out from the core. These are the colorful strings seen around the outside of the core, most readily visible in the ribbon diagrams in the two right panels. These are the N-terminal ends of all four histone polypeptides and, to a lesser extent, the C-termini of H2A and H2B. Positively charged amino acids (lysine and arginine) are over-represented in the tails, and this positive charge facilitates the interaction between negatively charged DNA and the histone octamer.
Test Your Understanding
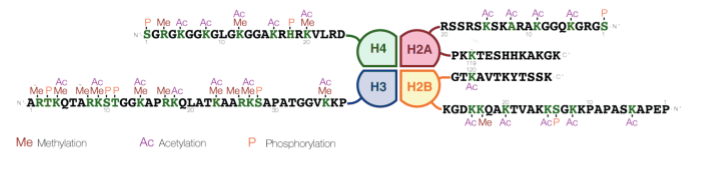
The histone tails can be modified through the covalent linkage of functional groups like phosphate, acetyl, and methyl groups to the side chains of particular amino acids. Most commonly, the positively charged lysines and arginines are targets for modification. Figure 4 illustrates the amino acids on each histone that can potentially be modified. This image uses the one-letter code for each amino acid, so lysine is indicated with a K and arginine with an R.[1] The modifications are sometimes called the histone code, signaling to the cell which parts of the genome should be transcribed. These changes in histone structure impact how tightly histones associate with the DNA, how densely chromatin is packaged within the nucleus, and the accessibility of the DNA for transcription and gene expression.
Differences in chromatin packaging can be seen within the nucleus using both light and electron microscopes. Densely packed chromatin is called heterochromatin. Epigenetically silenced regions of the genome are typically packed in heterochromatin, and heterochromatin has few actively transcribed genes. Euchromatin is more loosely packed, and most actively transcribed genes are found in euchromatic regions of the nucleus.
Media Attributions
- Nucleosome and chromatin structure © David O Morgan via. Wikimedia is licensed under a CC0 (Creative Commons Zero) license
- Three views of nucleosome © Darekk2 via. Wikipedia is licensed under a CC BY-SA (Attribution ShareAlike) license
- Histone modifications © Modified from Mariuswalker, Based on Rodriguez-Paredes and Esteller, Nature, 2011 via. Wikipedia is licensed under a CC BY-SA (Attribution ShareAlike) license
- Rodriguez-Paredes, M. & Esteller, M. Cancer epigenetics reaches mainstream oncology. Nat. Med. 17, 330 (2011). ↵

