Chromosome Structure
In eukaryotes, DNA is found in the nucleus as well as in the chloroplasts and mitochondria. In prokaryotes, the DNA is not enclosed in a membranous envelope. The cell’s entire genetic content (all its DNA) is its genome, and the study of genomes is genomics.
An organism’s genome is typically divided among a number of long DNA molecules called chromosomes. While prokaryotic genomes are typically made up of only one chromosome, eukaryotic genomes may have many. Human cells, for example, have 23 pairs of nuclear chromosomes (46 total) plus one mitochondrial chromosome.
Each chromosome can be thousands or even millions of base pairs long. The largest human chromosome (Chromosome 1) is 249 million base pairs long. As shorthand, biologists sometimes refer to lengths of DNA in kilobases or megabases, using the prefixes of the metric system. A thousand base pairs would be one kilobase, and human Chromosome 1 is 249 megabases. The human genome in total is 6 gigabases.
Prokaryotic chromosomes are typically circular, while eukaryotic chromosomes are typically linear in structure. In eukaryotic cells but not in prokaryotes, DNA forms a complex with histone proteins to form chromatin. The protein and DNA together make up the substance of eukaryotic chromosomes.
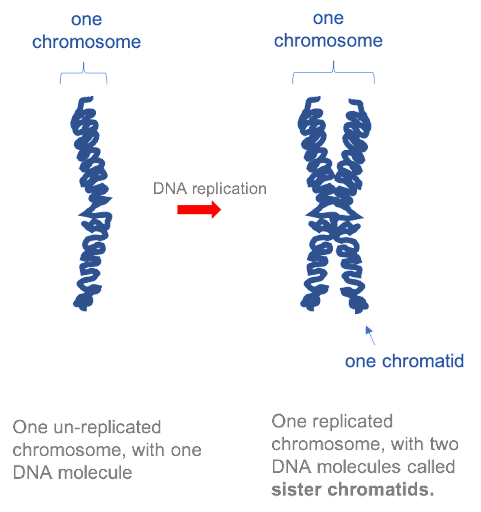
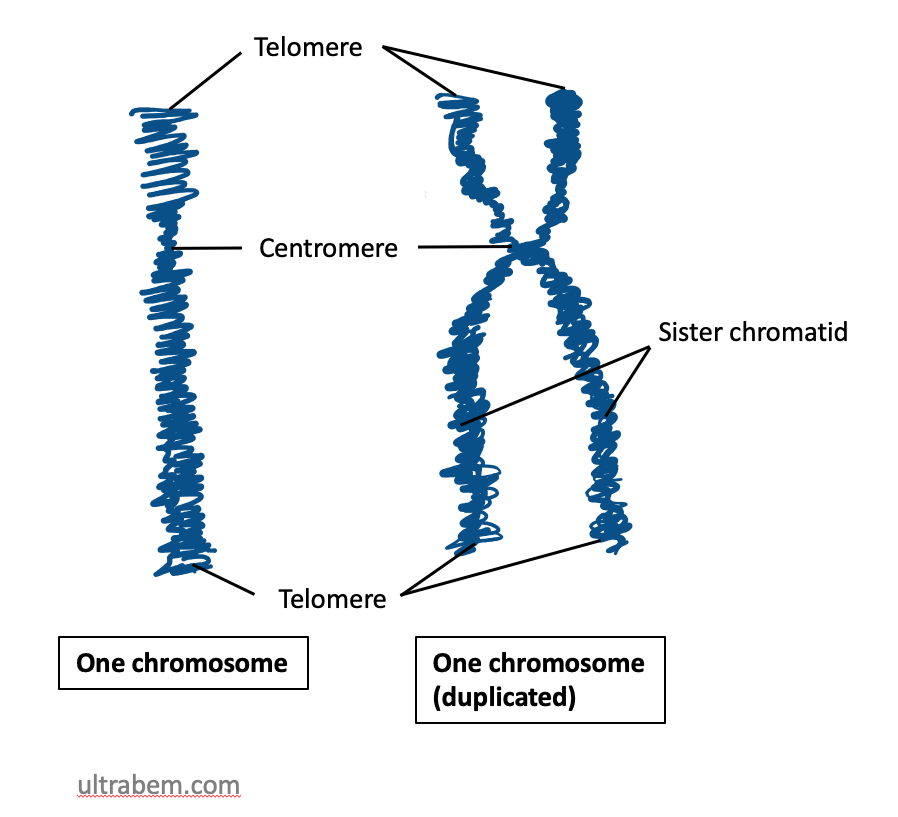
Before a cell can divide, the entire genome must be copied, so that there is a complete set of chromosomes available to be passed to each daughter cell. In the first half of the cell cycle, each eukaryotic chromosome is made up of one molecule. However, after each chromosome has been copied, the two daughter molecules remain linked together by specialized proteins. The two DNA molecules are called sister chromatids, and as long as they remain linked they are still collectively called one chromosome. This is illustrated in Figure 15. Note that although the replicated chromosome is drawn here (and in many other images!) as an X-shaped structure, in actuality the two chromatids are bound together all along their length. Individual chromatids cannot visually be distinguished using light microscopy, because they are so tightly associated with one another (see Figure 19 for an example).
A chromosome may contain tens of thousands of genes. Many genes contain the information to make protein products. Other genes code for RNA products. All cellular activities are ultimately controlled by turning the genes “on” or “off” – using the genetic information to make RNA and protein products, or not. Whether or not a gene is active can have profound effects on the function of a cell, allowing differences in cell behavior, appearance, and function.
DNA can be highly compacted
If stretched to its full length, the DNA molecule of the largest human chromosome would be 85mm. Yet during mitosis and meiosis, this DNA molecule is compacted into a chromosome approximately 5µm long. Although this compaction makes it easier to transport DNA within a dividing cell, it also makes DNA less accessible for other cellular functions such as DNA synthesis and transcription. Thus, chromosomes vary in how tightly DNA is packaged, depending on the stage of the cell cycle and also depending on the level of gene activity required in any particular region of the chromosome.
Prokaryotic compaction
Prokaryotic chromosomes are circular DNA, with one continuous circular sugar-phosphate backbone. They are typically smaller than eukaryotic chromosomes. For example, the smallest human chromosome, chromosome 21, is 48 million base pairs (48,000,000 base pairs, or 48 megabases). The largest human chromosome, chromosome 1, E coli, a bacterium commonly used as a model organism in the lab, has a genome comprised of one circular piece of DNA, about 4,600,000 base pairs (or 4.6 megabases).
4.6 megabases is still 1.5 millimeters long – 10x longer than the length of the E. coli cell, which is only 1-2 micrometers. To fit a long piece of DNA into a relatively small cell, prokaryotes typically compact their DNA through supercoiling. This results from over- or under-winding the DNA, which causes it to curl up or writhe around itself. This is like what happens if you try to untwist or overtwist a piece of string comprised of smaller individual twisted strands. As shown in the video in Figure 16, over- or under-twisting a coil introduces writhes in the circular structure, packing what was a loose, floppy structure into a tightly coiled package. Most prokaryotes compact their DNA through underwinding, although some extremophiles use overwinding.
Levels of compaction in eukaryotes
Eukaryotic chromosomes are linear, so writhes introduced by under-winding are not retained in the structure. Instead, eukaryotic cells use proteins to organize long, linear DNA molecules into compact structures. Collectively, the DNA and proteins together are called chromatin.
There are several different levels of structural organization in eukaryotic chromosomes, with each successive level contributing to the further compaction of DNA. Each level involves a specific set of proteins that associate with the DNA to compact it.
First, proteins called the core histones act as spool around which DNA is coiled twice to form a structure called the nucleosome. The core histones are called H2A, H2B, H3, and H4. Two of each core histone come together to form a histone octamer. The histones each have long tails extending from the core of the histone. These tails contain positively charged amino acids, which bind to the negatively charged DNA to hold it in place. About 150 bp of DNA wraps around each histone octamer, as shown in Figure 17.
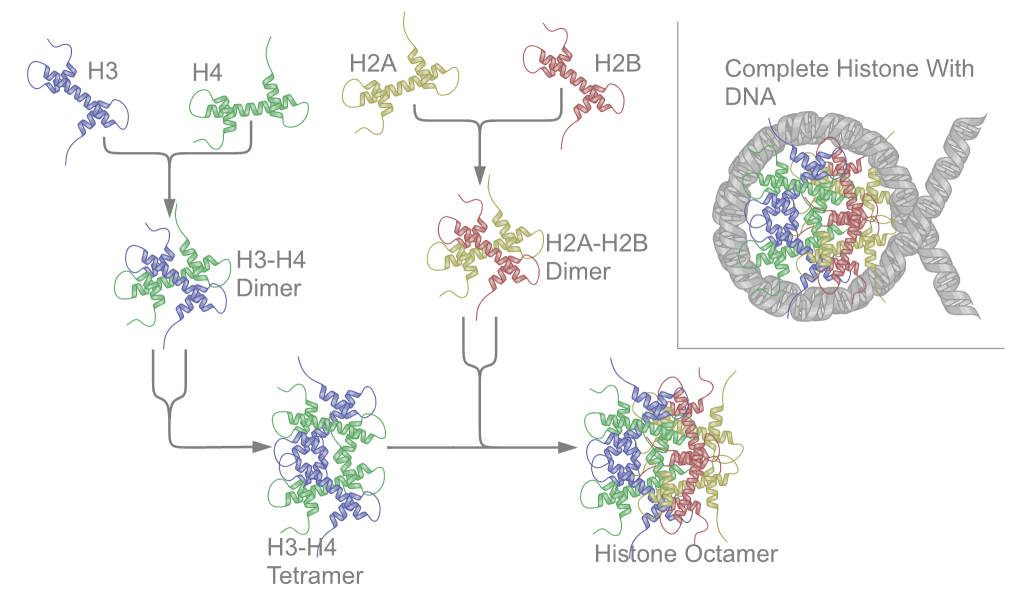
Nucleosomes are formed at regular intervals along the DNA strand, giving the molecule the appearance of “beads on a string” under an election microscope. At the next level of organization, histone H1 helps to compact the DNA strand and its nucleosomes into a 30nm fiber, so named because it is 30nm in diameter. Subsequent levels of organization involve the addition of scaffold proteins that wind the 30nm fiber into coils, which are in turn wound around other scaffold proteins (Figure 18).
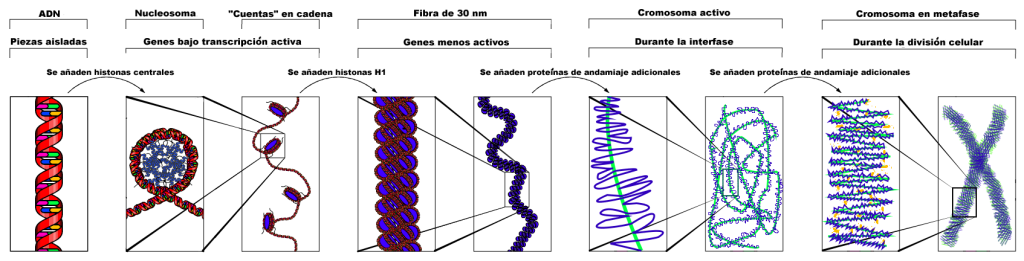
For more loosely compacted DNA, only the first few levels of organization may apply. DNA is fully compacted, or condensed, only during cell division. During interphase of the cell cycle, the less compact DNA is said to be decondensed. Note again: Although replicated chromosomes are often depicted as “X” shaped structures, as in this image, scaffold proteins bind the length of sister chromatids. Under a microscope, replicated chromosomes can appear as a single linear shape since the two chromatids may not be resolved.
Chromatin Packaging Varies inside the Nucleus: Euchromatin & Heterochromatin
Chromosomes stain with some types of dyes, which is how they got their name: Chromosome means “colored body”. Certain dyes stain some regions along a chromosome more intensely than others, giving some chromosomes a banded appearance. Researchers skilled in cytogenetics can use these characteristic banding patterns to identify specific chromosomes. This is discussed further in later modules.
There are two major types of chromatin, but these are more the ends of a continuous and varied spectrum. Euchromatin is more loosely packed, and tends to contain genes that are being transcribed. Heterochromatin is more densely compacted. It tends to contain many repetitive sequences and genes within heterochromatin tend not to be transcribed.
Morphological features of Chromosomes
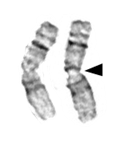
Chromosomes also contain distinctive features, such as centromeres and telomeres. Both of these are usually heterochromatin. In most cases, each chromosome contains one centromere. Centromeres are regions of the chromosome that are bound by proteins that link the centromere to microtubules that transport chromosomes during cell division. Under the microscope, centromeres of metaphase chromosomes can sometimes appear as constrictions in the body of the chromosome (Figure 19).
If a centromere is located near the middle of a chromosome, it is said to be metacentric, while an acrocentric centromere is closer to one end of a chromosome, and a telocentric chromsome is at, or near, the very end. In contrast, some species have a holocentric centromere, where no single centromere can be defined and the entire chromosome acts as the centromere.
Telomeres are repetitive sequences near the ends of linear chromosomes and are important in maintaining the length of the chromosomes during replication, and protecting the ends of the chromosomes from alterations.
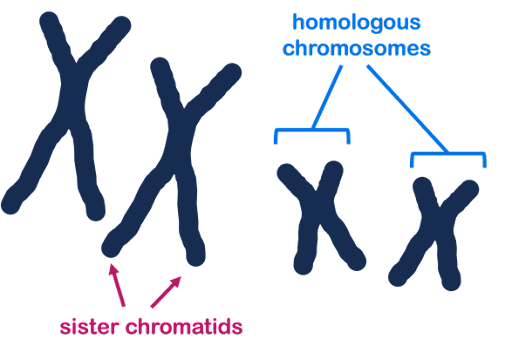
Some chromosome terminology: Homologous is a word that means similar but not identical. Homologous chromosomes are pairs of similar but non-identical chromosomes in which one member of the pair typically comes from the male parent, and the other comes from the female parent. Homologous chromosomes contain the same genes but not necessarily the same version (allele) of each gene. In Figure 20, three chromosomes are shown: two homologous large chromosomes and one smaller chromosome that is not homologous to the other two.
Sister chromatids are the two DNA molecules that form halves of a single, replicated chromosome. In Figure 20, each chromosome has two sister chromatids.
Because a pair of sister chromatids is produced by the replication of a single DNA molecule, their sequences are essentially identical (same versions of each gene), differing only because of any DNA replication errors. Homologous chromosomes will be highly similar, but not identical. They may have different versions (alleles) of each gene.
Nuclear organization
Even when they are decondensed, the chromosomes are not randomly arranged in within the interphase nucleus. They often have specific locations within the nucleus and relative to one another. In Figure 21, each chromosome in the nucleus of a single cell has been stained a different color, using a technique called Fluorescent In Situ Hybridization (FISH). FISH is sometimes called chromosome painting, since it is so colorful! Rather than decondensed chromosomes tangling all over the nucleus like a big plate of spaghetti, each chromosome remains collected in a small area of the nucleus. As we will see in other modules, this likely helps in gene regulation.
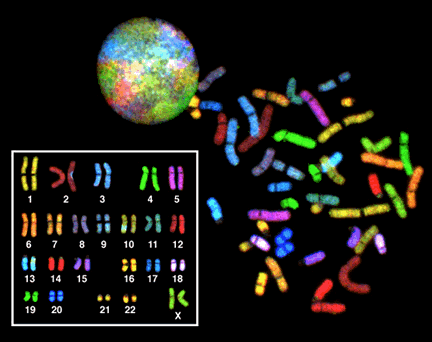
Media Attributions
- NA replication © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license
- Nucleosome structure © Wikimedia is licensed under a CC0 (Creative Commons Zero) license
- Estructuras cromatina © Wikipedia is licensed under a CC0 (Creative Commons Zero) license
- metacentric chromosomes © LibreText is licensed under a CC0 (Creative Commons Zero) license
- Chromosome structure © wikipedia is licensed under a CC0 (Creative Commons Zero) license
- chromosomes and chromatids © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license
- FISH (Fluorescence in situ hybridization) © Wikipedia is licensed under a Public Domain license

