Complementation
As discussed in the Basic Heredity module, recessive phenotypes are always “true-breeding”: a cross between two individuals, both with a recessive phenotype, will always have recessive offspring. But in some cases, complementary genes give rise to a situation that appears to violate this principle. This happens when mutations in two different genes can produce similar phenotypes. Such genes are sometimes called complementary genes.
Because complementary genes give the same phenotype when mutated, complementary genes give a 9:7 phenotypic ratio in a dihybrid cross (Table 2). Complementary genes often participate in the same biochemical pathway: they give a 9:7 ratio in a dihybrid cross because a loss of function of either of the genes would block the function of the whole pathway. Several examples of how complementary genes might work together to perform a biochemical function – the production of purple pigment – are shown in Figure 15.
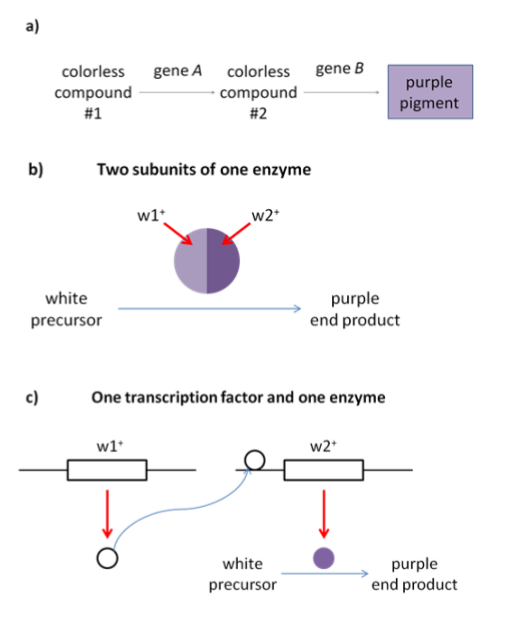
A simple graphic showing one transcription factor and one enzyme resulting in purple color
The word complementary comes from the word “complement”, which shares a root word with “complete”. (Not “compliment”, which means to say nice things.) You can think of complementary genes as forming a complete pathway. Without the complete whole, the pathway won’t function, so a homozygous loss of function of any gene will give a mutant phenotype.
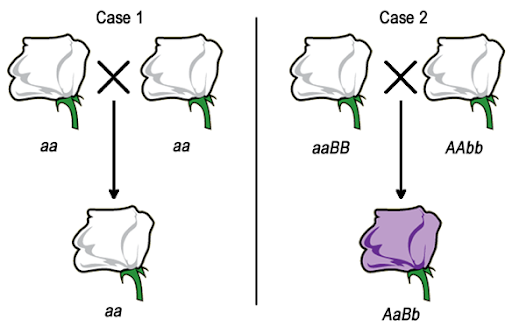
Consider a hypothetical situation where purple flowers are the wild type, and white flowers are a recessive mutation. Because white flowers are recessive, we’d expect a cross between any two white flowers to produce white offspring, as shown in Case 1 in Figure 16. But if the white color is caused by recessive mutations in different genes in each parent, the dihybrid offspring will have a wild type phenotype.
An example of this in humans: a number of genes contribute to a recessive phenotype of deafness. The phenotype requires homozygosity for the deafness-associated alleles, so we might therefore expect that two Deaf parents would always have a Deaf child (aa x aa). However, in some cases two Deaf parents may have a hearing child. This happens when the parents are homozygous for deafness-associated alleles of different genes (AAbb x aaBB).
How many genes? Complementation tests
Biochemical pathways can get pretty complicated. In the previous examples, we gave examples where two genes are controlling the same phenotype, but most biochemical pathways involve far more than just two genes.
A complementation test can be used to figure out if two related traits (like the hypothetical situation of purple vs white flowers, for example) are a result of different alleles or different genes. Complementation tests can also be used to figure out how many genes are contributing to a particular trait.
In its simplest form, a complementation test is a cross between two individuals with the same recessive phenotype. The F1 offspring indicate whether the parents have mutations in the same gene or mutations in two different genes. As shown in Figure 16, there are two possibilities for the results of a complementation test. If two white parental flowers produce white offspring, we say that the parental mutations fail to complement one another. The F1 offspring does not have a complete set of the wild-type alleles needed to produce pigment. But if two white flowers are crossed to produce wild type (purple) offspring, the parental mutations complement one another. The F1 offspring must have a complete set of the alleles needed to produce the wild type phenotype.
In the previous example, we considered a situation where two genes controlled the production of purple pigment. But what if there were more? Multiple pair-wise crossings can give geneticists a clue as to how many genes might be controlling a phenotype.
Consider a situation where there are 6 true-breeding lines of white flowering plants. How can we use a complementation test to determine how many genes are affecting flower color? We can perform multiple crosses, crossing every line with every other line, one pairing at a time. The results of such a test are shown in Table 4. The boxes of the table list the phenotype of the F1 offspring for each of the indicated crosses. For example, when strain 1 is crossed with strain 2, the offspring are wild type (purple), but when strain 1 is self-crossed the offspring are white. The empty/dark gray boxes in the table are crosses that are represented elsewhere in the table: the cross between strains 1 and 2 is in the first column second row, but the first row second column is empty.
| Strain | 1 | 2 | 3 | 4 | 5 | 6 |
| 1 | Mutant | No Data | No Data | No Data | No Data | No Data |
|---|---|---|---|---|---|---|
| 2 | Wild-type | Mutant | No Data | No Data | No Data | No Data |
| 3 | Wild-type | Wild-type | Mutant | No Data | No Data | No Data |
| 4 | Wild-type | Wild-type | Mutant | Mutant | No Data | No Data |
| 5 | Mutant | Wild-type | Wild-type | Wild-type | Mutant | No Data |
| 6 | Mutant | Wild-type | Wild-type | Wild-type | Mutant | Mutant |
For the example shown in Table 4, you’ll see that all the self-crosses listed (1×1, 2×2, 3×3, etc) give a mutant phenotype. This is what you would expect from a true-breeding strain! Complementation tests will usually include the self-cross offspring as a control.
If you look at all of the crosses involving strain 1, you’ll see that crosses with strains 2, 3, and 4 produce wild-type offspring. This means that strain 1 complements strains 2, 3, and 4. Those strains have a different mutation than strain 1.
But strains 5 and 6 do not complement strain 1: offspring of those crosses have a mutant phenotype. From this, we say that strains 1, 5, and 6 belong to the same complementation group. Members of a complementation group are presumed to have mutations in the same gene.
Can you find a set of two more genes that do not complement? Answer: Strains 3 and 4 also belong to the same complementation group.
So which strain is left? Strain 2 complements all other strains, so we say that strain 2 is in a “group” by itself. That gives us three groups:
- Group A: Strains 1, 5, 6
- Group B: Strains 3, 4
- Group C: Strain 2
We started with 6 strains, and all strains must be assigned to one group. Because there are three complementation groups, we can conclude that there are likely to be at least three genes controlling flower pigmentation. Why the at least? If we expanded our sample size to include additional strains, we might find additional strains that belong to each group, or we might discover additional complementation groups.
Test Your Understanding

What about a situation where there are more mutant phenotypes? Well, a complementation test works here, too. Flowers come in more than just two colors, and remember, there may be more than two alleles of a gene in a population. Let’s look at a hypothetical situation where we have flowers with multiple mutant phenotypes.
In a complementation test for the four mutant strains, we see the results shown in Table 5.
| Strain | 1 | 2 | 3 | 4 |
| 1 | White | No Data | No Data | No Data |
|---|---|---|---|---|
| 2 | Wild type | White | No Data | No Data |
| 3 | Wild type | Blue | Blue | No Data |
| 4 | White | Wild type | Wild type | White |
As with the simpler two-phenotype example, here we look for complementation (wild type offspring) and failure to complement (mutant offspring). The cross between strains 2 and 3 fails to complement since the offspring have a mutant phenotype (blue). From this, we can conclude that strains 2 and 3 have mutations in the same gene. Further, because strains 2 and 3 have different phenotypes, we conclude that strains 2 and 3 have two different alleles of the mutant gene. We also can conclude that blue is dominant to white in the allelic series.
Test Your Understanding
This example also reveals something important about complementation groups: although every member of the group harbors a mutation in the same gene, they do not all share the same mutant allele. As an example, the white gene in Drosophila has over 300 known mutant alleles. All of these alleles would be part of the same complementation group.
Limits of the complementation test
The complementation test is limited in that it will only work when the mutant phenotype is recessive and if each tested individual has a mutation in one gene. In some cases, similar phenotypes may be caused by a dominant mutation in one gene or a recessive mutation in another. In such cases, the F1 generation of a cross with the dominant mutation will always include mutant individuals, regardless of the genotype of the other parent. This may be easily confused with a failure to complement, so care must be taken in interpreting results of a complementation test. Results also can be complicated if a mutant strain has a mutation in more than one gene.
Variations of the complementation test in human biology
A complementation group refers to a group of alleles of a single gene that contributes to a complex, multigenic phenotype. The members of a complementation group will always fail to complement each other, since they are all missing the ability to perform the same biological function. Many complex human phenotypes are grouped in complementation groups, even though the traditional complementation test described above cannot be used in humans.
But many complex human traits also confer a cellular phenotype that can be measured in the lab. Cellular and molecular genetics techniques can be used to determine if the phenotype of a patient’s cells can be complemented. An example of this is Fanconi anemia. Patients with Fanconi anemia are usually diagnosed in childhood with bone marrow failure. They are highly susceptible to certain forms of cancer, and they may have characteristic skeletal abnormalities, hyper-pigmented spots on the skin, and/or small stature. There is no cure for the disease, although many patients are ultimately treated with bone marrow transplant.
As of the writing of this text, there are 22 known Fanconi anemia complementation groups[1]. The groups are named for the disease followed by a letter: FancA, FancB, FancC… all the way to FancY. Each group corresponds to a particular gene. The Fanconi genes encode proteins that participate in the repair of a particular form of DNA damage: inter-strand crosslinks. An inter-strand crosslink is a covalent bond that links the two complementary strands of the double helix in a way that prevents them from separating during replication or transcription.
Healthy cells constantly sustain DNA damage, but cells have repair mechanisms built in place to fix the damage before DNA is replicated and/or the cell divides. Different types of damage are repaired with a different set of repair enzymes, so Fanconi cells can repair some damage but not inter-strand crosslinks.
Cells collected from Fanconi anemia patients are highly susceptible to DNA crosslinking agents since they cannot repair the damage. In fact, this is how patients are often diagnosed: blood or skin cells isolated from a patient are treated with a DNA crosslinking drug. If a patient’s cells are killed by the DNA crosslinks but not other forms of DNA damage, it is an indication of Fanconi anemia. This is also the basis for a version of a complementation test that, until recently, was the most common way to determine which FA-associated gene was affected in each patient.
Cells from a newly diagnosed Fanconi anemia patient would be fused with a panel of cells from known FA complementation groups. The fused cell (now with four copies of each chromosome) would be treated with DNA crosslinking agents. If the fused cells die, the patient’s cells do not complement the test cells, indicating the patient’s cells have a loss of function pathogenic variant in the same gene as the test cell line. If the fused cells survive, this indicates the patient’s cells complement the test cells, and the patient has a pathogenic allele in different gene from the test cell line. This is important in treating the symptoms of Fanconi anemia, since patients belonging to different complementation groups may have slightly different disease phenotypes and react differently to treatment.
If a new complementation group was identified, a related experiment could then be used to identify the genomic locus responsible for the FA phenotype. Genomic DNA from a healthy (non-FA) individual is broken into thousands of fragments and inserted into small, circular DNA molecules called plasmids. The plasmids carrying the genomic segments are introduced into FA cells, one segment per cell. The cells are then subjected to DNA crosslinking agents. Cells that survive have been complemented by the genomic segment they host. The genomic DNA is recovered from the cells for further study, thus identifying the gene linked with FA.
This process revealed that many of the genes represented by the complementation groups were already known by other names, studied by molecular geneticists in other contexts. For example, FANCD2 is the BRCA2 gene. Some mutations in the BRCA2 gene are associated with a predisposition to breast cancer, while other mutations in the BRCA2 gene are associated with Fanconi anemia. (Most people have a healthy version of the gene and have neither Fanconi anemia nor a genetic predisposition to breast cancer.)
Both of these techniques have largely been superseded by newer diagnostic tests, including whole-genome sequencing. But the vocabulary of complementation remains: like many multigenic disorders, Fanconi anemia is still subdivided into complementation groups, and as new Fanconi-associated mutations are identified through whole-genome sequencing, new complementation groups are added to the list.
Media Attributions
- Molecular mechanisms © Open Genetics is licensed under a CC BY-NC-SA (Attribution NonCommercial ShareAlike) license
- Cross between two white flowers © Original-Deyholo via. Open Genetics Lectures 2017 is licensed under a CC BY-NC (Attribution NonCommercial) license
- Flowers © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license
- Mehta, P. A. & Ebens, C. Fanconi Anemia. in GeneReviews® (eds. Adam, M. P. et al.) (University of Washington, Seattle, Seattle (WA), 1993). ↵

