Enhancers in action: Drosophila development and the eve gene
A classic example of eukaryotic gene regulation in action is seen during early development of the Drosophila melanogaster embryo. The eve gene described in the earlier section on enhancers is part of this process. The eve gene was first identified by Wieschaus and Nüsslein-Volhard in a search for embryonic mutants.
About 24 hours after fertilization of the egg, a wild type Drosophila embryo looks like Figure 13 with visible segments. In this image, the anterior (head) part of the embryo is to the left, and the posterior (tail) is to the right.
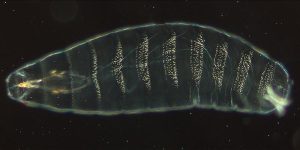
Wieschaus and Nüsslein-Volhard searched for misshapen, mutant embryos that looked different to the wild type. Exposure to mutagens (chemicals that introduce mutations into DNA) leads to in increased frequency of mutant phenotypes. This type of experiment is called a forward genetic screen, where mutant embryos are screened for interesting phenotypes. See Figure 14 for a comparison of forward and reverse genetic screens.
Nüsslein-Volhard and Wieschaus searched painstakingly through mutant Drosophila embryos, looking for those with mutations affecting development. And they did find misshapen embryos! Most of these embryonic phenotypes were lethal, so the embryo could not grow into an adult fly. But the early embryos had clearly visible mutant phenotypes, with the wild-type pattern of segmentation disrupted. You can see images of the mutant phenotypes in their 1980 paper.
Some mutant embryos were missing multiple segments in the middle of the embryo. Others were missing every other segment or showed structures reversed in orientation. (Yes, they looked very weird.) The eve mutants were missing every even-numbered segment, so the gene was named even-skipped, or eve for short.
Studies of these embryonic mutants revealed the underlying genetic rules that control development in Drosophila. Eric Wieschaus, Christiane Nüsslein-Volhard, and Edward Lewis were awarded the Nobel Prize in Physiology or Medicine in 1995 for this work.
Note: Although the examples given here are from Drosophila, these genetic mechanisms were later shown to be conserved in other organisms. Today, we know that human versions of genes important for patterning in the Drosophila embryo also are important in human development. The regulation of those genes is what makes it possible to build a multicellular organism with differentiated tissues.
Test Your Understanding
Eve expression during development
For multicellular eukaryotic organisms that reproduce sexually, all cells of the body originate from a single cell: the zygote is the fertilized egg that contains the genetic information from both parents. The zygote undergoes subsequent rounds of cell division as it develops into an embryo. As the cells divide, they become specialized so that different tissues can perform different tasks within the body. This process of specialization is called differentiation. At each step of the differentiation process, changes in gene expression cause a different set of proteins to be produced, which affects cell phenotype. To use vocabulary of other modules, each cell of a developing embryo has the same genome but a different transcriptome and proteome.
Remember from earlier in this chapter: the gene eve is expressed in multiple stripes along the length of an embryo. Embryos mutant for eve are missing every other segment – the ones where eve is normally expressed. Expression of eve in this stripe-like pattern is controlled by five enhancers, as shown in the top panel of Figure 15.
Each of the five eve enhancers has binding sites for multiple factors. The stripe 2 enhancer is diagrammed in Figure 15.
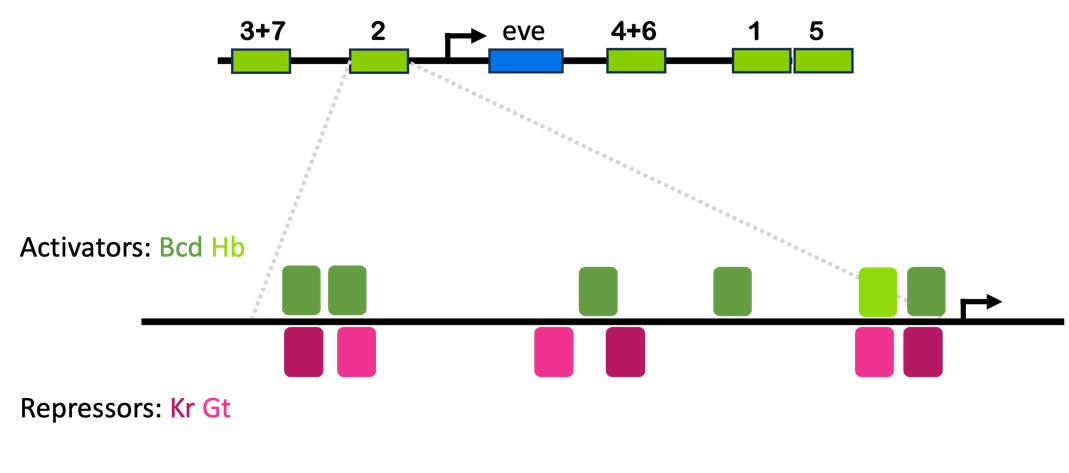
The stripe 2 enhancer drawn in Figure 15 is about 500 base pairs long. It has binding sites for activators Bicoid (Bcd, dark green) and Hunchback (Hb, light green), as well as repressors Giant (Gt, light pink) and Kruppel (Kr, dark pink)8. These funny-named factors are all named for the phenotypes observed in mutant embryos – more on that later.
You’ll notice that the activator binding sites overlap with many of the repressor binding sites. If repressors are bound to these elements, activators presumably cannot bind. In this way, the enhancer drives expression of the eve gene in any tissue that has both activators (Bcd and Hb) but neither of the repressors (Gt and Kr).
This is important: it allows the eve gene to be turned on, but also to be turned off! Proper gene regulation means that a gene is turned on when appropriate and turned off when appropriate. Not all genes are expressed in all tissues!
Bicoid, Hunchback, Giant, and Kruppel are expressed in varying patterns along the length of the Drosophila embryo, from anterior to posterior, as shown in Figure 16. About one third of the way from the anterior of the embryo, there is one area several cells wide where both Bicoid and Hunchback proteins are present and where Kruppel and Giant proteins are not. This is where eve stripe 2 is expressed.
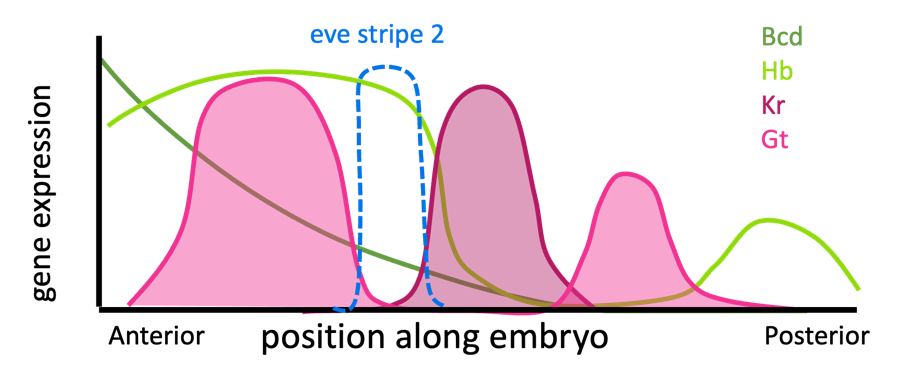
You can think of Bicoid and Hunchback as activating eve expression everywhere in the anterior end of the embryo, but Giant and Kruppel block the edges of that expression. (Note: Additional influences not discussed here limit expression of eve at the anterior-most end of the embryo.)
The four other enhancers are regulated by these factors and others, with each enhancer acting as its own independent switch. For example, the stripe 3+7 enhancer is activated by proteins found everywhere in the embryo, but it is inhibited by Hunchback and a protein called Knirps, as shown in Figure 17. (Knirps is pronounced “nerps”).
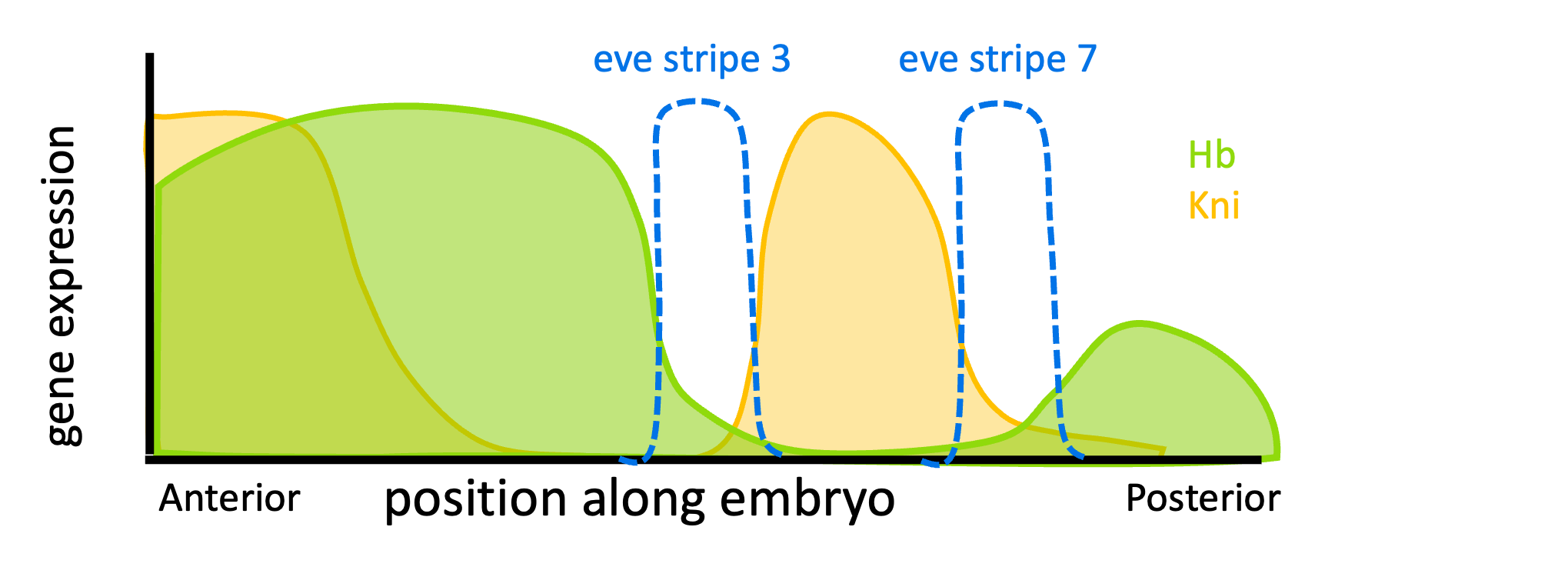
Because of the different arrangements of binding sites, Hunchback acts as an activator of stripe 2, but a repressor on stripes 3 and 7! Many transcription factors act to both activate and repress transcription, depending on context. This is true in both prokaryotes and eukaryotes – the lambda repressor described in the Overview of Gene Regulation chapter was another example.
Note that only one enhancer needs to be active for the gene to be expressed. Conditions are right for the stripe 3+7 enhancer to drive eve expression in stripes 3 and 7 of the embryo. Conditions are right for the stripe 2 enhancer to be active in stripe 2. The same is true for other enhancers in other regions of the embryo.
Test Your Understanding
A note on Drosophila naming conventions
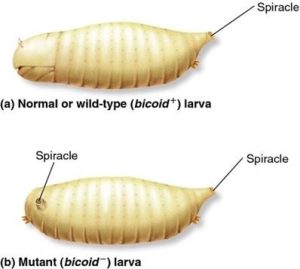
Most of these genes were initially discovered in forward genetic screens for homeotic or embryonic mutants, and the names that reflect the original researcher’s description of the mutant phenotype. For example, the name bicoid is derived from the fact that mutant embryos lacked anterior (head) structures. These mutants were described as “bicaudal”, which means having two tails. An illustration of a bicoid mutant embryo is shown in Figure 18, and you can see a micrograph of a bicoid mutant embryo at the CSH DNA Learning Center. Some of the names are quite whimsical: the name gooseberry reflects the round mutant embryo that sort of resembled a gooseberry (Figure 19), and armadillo mutants resembled a little armadillo.

It should be acknowledged, though, that some of the names reflect their times: Kruppel, for example, is derived from a German word meaning cripple, and hunchback was named for the shape of the unusual shape and distribution of denticles. A fruit fly doesn’t care if it’s called a cripple, but this becomes relevant to people because many of these genes have human orthologs that have implications for human health and disease. A gene name like “lunatic fringe” or “Indy” (“I’m not dead yet”) or “roadkill” can make even more difficult conversations with patients who are already facing challenges.
In the chapter on Mutation, we discussed the rationale for a shift from the word “mutant” to the word “variant” when describing human genetic variation. Here is another example of how terminology has changed over time as scientists and medical professionals recognize the impact of the vocabulary. The human versions of some of these genes have been renamed,[1] often as abbreviations that become the more common nomenclature (e.g. Kruppel-like Factor 1 is KLF1 ).
Media Attributions
- Figure 2 EG Regulation is licensed under a Public Domain license
- Figure 14 EG Regulation © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license
- Figure 15 EG Regulation © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license
- Figure 16 EG Regulation © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license
- Figure 17 EG Regulation © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license
- Figure 18 EG Regulation © Charodeika ya is licensed under a CC BY-SA (Attribution ShareAlike) license
- Figure 19 EG Regulation © neurovelho is licensed under a CC BY-SA (Attribution ShareAlike) license
- Hopkin, M. Troublesome gene names get the boot. Nature news061106-2 (2006) doi:10.1038/news061106-2. ↵

