Eukaryotic transcription
In prokaryotes, there is a single RNA polymerase responsible for transcription of all genes. But eukaryotes have three RNA polymerases. RNA Polymerase I (RNA Pol I) transcribes most rRNA; RNA Pol II transcribes mRNA; RNA Pol III transcribes tRNA. Pol II and Pol III each transcribe some snRNA and miRNA. Many of these RNA molecules are also processed post-transcriptionally before they become active.
Test Your Understanding
Like the prokaryotic RNA polymerase, all three eukaryotic polymerases require DNA elements to recognize a promoter, synthesize the transcript, and release the RNA from the transcription machinery. But the prokaryotic RNA polymerase holoenzyme including sigma factor can recognize promoters and begin transcription without other cofactors. The eukaryotic polymerases cannot: they require the action of additional general and regulatory transcription factors to transcribe. Because of this, eukaryotic promoters tend to be more complicated than bacterial promoters.
Although there are similarities in the mechanism of action for all three eukaryotic polymerases, each recognizes a different type of promoter, and each interacts with its own set of transcription factors.
RNA polymerase II transcription initiation
Let’s look in detail at RNA Pol II transcription of mRNAs, the protein-coding RNAs. RNA Pol II requires the action of general transcription factors to initiate transcription. Most general transcription factors are named by convention TFII_, where TFII indicates they are transcription factors for RNA Pol II, and “_” is a letter that specifies individual factors. Some examples are TFIIB, TFIID, and TFIIH. (The other RNA polymerases also have designated transcription factors. Theirs are named TFI_ and TFIII_.)
Collectively, the general transcription factors and RNA polymerase are called the core or basal transcription machinery. The word basal has the same root word as “basic” or “basis”: it is the basic transcription machinery for all RNA Pol II-transcribed genes.
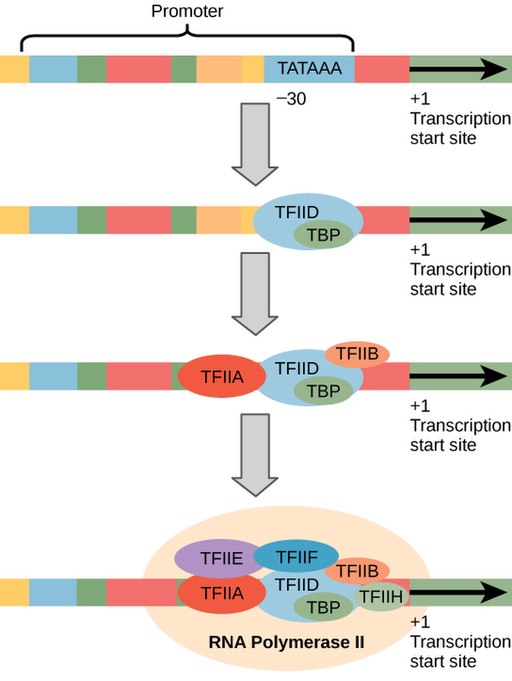
The general transcription factors assemble onto a core promoter about 30 base pairs from the start site of transcription. RNA Pol II-transcribed genes have a common sequence called the TATA box within the core promoter. The TATA box is so named because it has lots of T’s and A’s: the consensus sequence for the TATA box is “TATA(T/A)A(T/A)”, although individual genes vary slightly. Like the sequences of the -10 and -35 boxes, the consensus sequence is found in the nontemplate strand upstream of the transcriptional start site.
The TATA box serves a similar purpose as the -10 box in prokaryotes. It is bound by a protein called TATA binding protein (TBP). TBP is part of a larger complex called TFIID. Additional general transcription factors bind after TFIID, recruiting RNA Pol II to the promoter (Figure 9)
In addition to the TATA box, the promoter often contains other common sequence elements. Two such elements are GC boxes and the CAAT box, again named for the nucleotides of the consensus sequence. The CAAT box is usually found about 60-100 bases from the +1 site, and GC boxes about 80-110 bases from the +1 site. Eukaryotic promoters are highly variable! Not all genes have CAAT boxes or GC boxes. In fact, some genes don’t have TATA boxes, either! In some TATA-less promoters, the presence of multiple GC boxes is sufficient to assemble the transcription machinery and initiate transcription.
About 100-250 base pairs from the +1 site of transcription are additional proximal promoter elements that play a role in gene regulation: they may be bound by general transcription factors or regulatory transcription factors. Regulatory transcription factors are also called specific transcription factors: they are specific to the adjacent gene, in contrast to the general transcription factors that facilitate transcription of all RNA Pol II-transcribed genes. Regulatory elements and regulatory transcription factors are discussed more in detail in the module on Gene Regulation.
Thousands of base pairs away – upstream or even downstream! – eukaryotic promoters may have enhancers or silencers: DNA elements with binding sites for multiple regulatory transcription factors. The core promoter, proximal elements, and far-away enhancer/silencer elements are shown in Figure 10.
These distal regulatory elements help the polymerase bind to the promoter when conditions are right for the gene to be active. Although they can be tens of thousands of base pairs away from the core promoter when measured linearly, DNA is a flexible molecule! It bends, twists, and loops in the nucleus of the cell. Elements that are far apart on the chromosome can actually be adjacent in three-dimensional space if the DNA loops around.
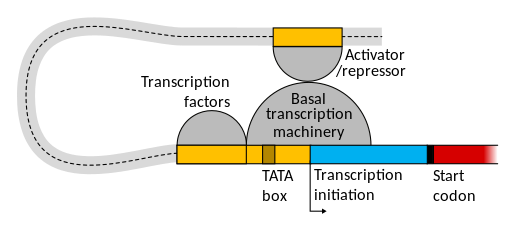
This is shown in Figure 11. Generally the proteins that bind to enhancers interact with other parts of the transcription machinery to help stabilize the polymerase at the promoter enough to initiate transcription. Like the proximal regulatory elements, enhancer elements will be discussed in more detail in the module on Gene Regulation.
RNA polymerase II termination
Once the polymerase and supporting transcription factors have bound to the promoter, the polymerase will synthesize the first nucleotides of the RNA, beginning at the +1 site and continuing downstream from the +1 site. But there are no terminators like those seen in prokaryotes. Instead, each of the three eukaryotic RNA polymerases terminates transcription via different DNA elements and/or protein factors.
RNA Pol II-transcribed genes are terminated via polyadenylation. Polyadenylation adds up to 250 adenosine nucleotides to the 3’ end of an mRNA molecule. In the process, transcription is terminated. These A nucleotides are not templated: there are no corresponding T’s in the template strand of the DNA. The length of A’s is called the poly-A tail.
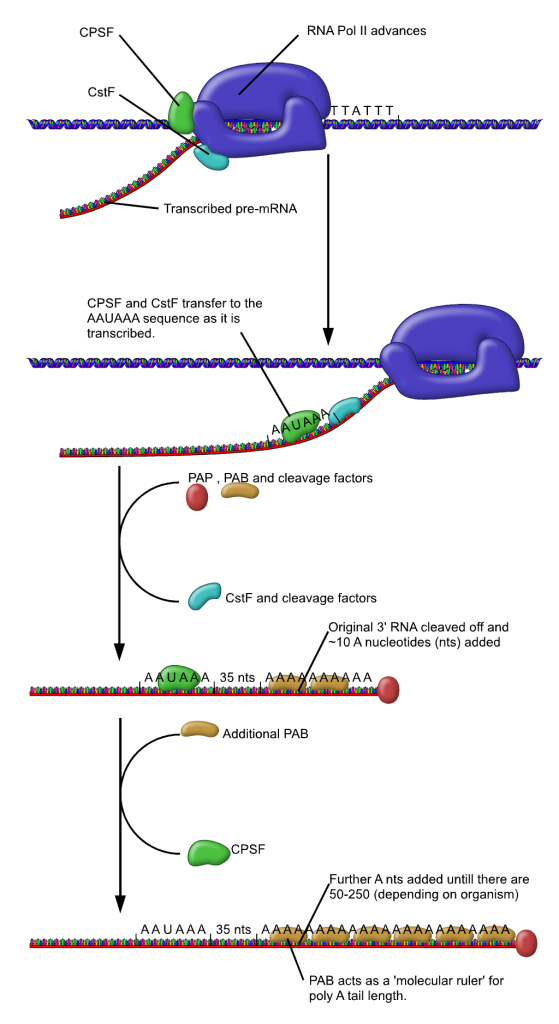
RNA pol II transcription machinery includes polyadenylation factors CPSF (Cleavage and Polyadenylation Specificity Factor) and CstF (Cleavage Stimulating Factor), which ride along with the polymerase as it transcribes. Once the polyadenylation signal sequence AAUAAA is transcribed, CPSF and CstF are transferred to the RNA molecule. CstF cleaves, or cuts, the RNA molecule about 35 nucleotides downstream of the polyadenylation signal, even while transcription continues at the 3’ end of the RNA. PAP, or Poly-A Polymerase, adds adenosine nucleotides to the new 3’ end of the cut RNA. This is shown on the left side of Figure 12.
Transcription of the original RNA molecule continues beyond the polyA signal sequence. Rat1 “torpedo” exonuclease then binds to the new 5’ end created by the cleavage. An exonuclease is an enzyme that breaks down nucleic acids starting at one end. Rat1 breaks down the remnant RNA from 5’ toward the 3’ end. It is called a “torpedo” exonuclease because one hypothesis is that this movement toward the 3’ end acts like a torpedo to break apart the DNA/polymerase complex and terminate transcription. This is shown on the right side of Figure 12. The dismantling of the transcription machinery is not well understood and likely requires the action of additional DNA elements and protein factors[1].
The transcripts of the other RNA polymerases, other methods of termination, and transcripts of RNA Pol I and RNA Pol III are not typically polyadenylated[2]. In addition to participating in transcriptional termination, Poly-A tails are important for the export of mRNAs from the nucleus and to the stability of mRNAs over time.
Test Your Understanding
Media Attributions
- Core promoter © Wikipedia is licensed under a CC0 (Creative Commons Zero) license
- Transcription initiation eukaryote B © Wikipedia is licensed under a CC0 (Creative Commons Zero) license
- Polyadenylation © Wikipedia is licensed under a CC0 (Creative Commons Zero) license
- Rodríguez-Molina, J. B., West, S. & Passmore, L. A. Knowing when to stop: Transcription termination on protein-coding genes by eukaryotic RNAPII. Mol. Cell 83, 404–415 (2023). ↵
- Arimbasseri, A. G., Rijal, K. & Maraia, R. J. Transcription termination by the eukaryotic RNA polymerase III. Biochim. Biophys. Acta 1829, 318–330 (2013). ↵

