Factors that influence cancer incidence: lessons from evolution
As mentioned at the beginning of this chapter, humans are not the only organisms to get cancer. It turns out that we can learn a lot from other organisms.
The effect of DNA damage is cumulative over time as lesions escape repair and become ensconced in the genome as mutations. And the more cells (and cell divisions) there are, one might expect, the more likely that one will be damaged in exactly the right way.
Within a species, this holds true: older organisms are far more likely to get most cancers than young ones, simply because it takes time to accumulate the mutations. The exceptions – childhood cancers like Rb or cancers that show up in young adulthood – appear to require fewer overall “hits”. Retinoblastoma, for example, is a two-hit cancer (one in each copy of the Rb gene), and CML appears to be a one-hit cancer (driven by the BCR-ABL oncogene).
But when we compare cancer across species, this doesn’t hold true. We might expect that larger and long-lived organisms, having more cells and presumably equally prone to DNA damage, would get cancer more frequently, and small shorter-lived organisms would get cancer more infrequently.
But that’s not the case. For example, elephants are large, long-lived organisms that rarely get cancer. So what’s going on?
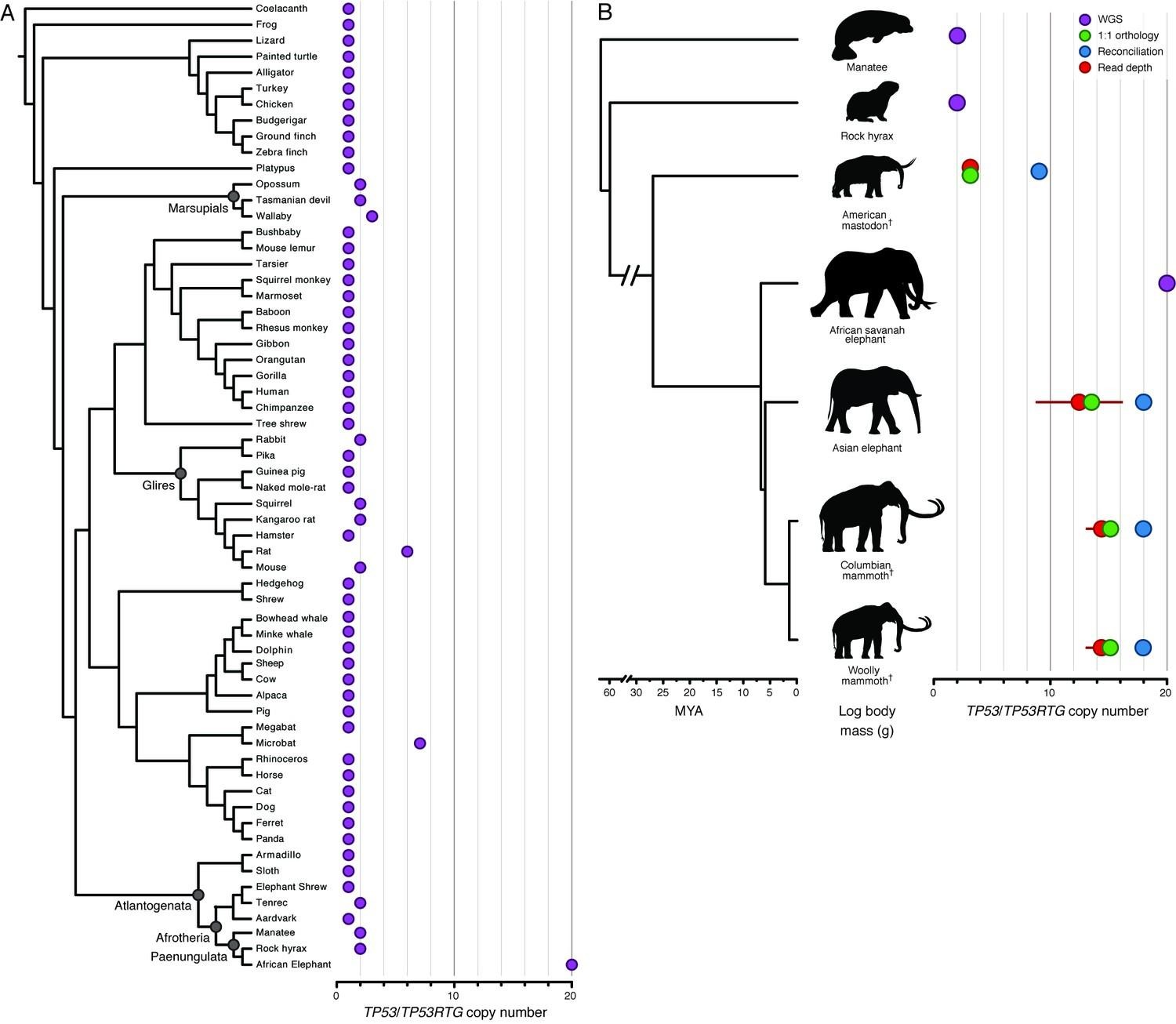
The answer is that body size in elephants likely co-evolved with additional anti-cancer defense mechanisms. Elephant genomes have multiple copies of the genes similar to the tumor suppressor protein p53, called TP53RTGs. Essentially, the p53 gene was duplicated multiple times over the modern elephant’s evolutionary history. More tumor suppressor genes = more tumor suppressor proteins = more tumor suppression!
In Figure 23, you can see a comparison of the number of copies of the p53 and related genes in a number of living and extinct proboscidean (elephant) species, arranged by evolutionary relationship. The African savannah elephant, the largest of the bunch, has the highest number of copies – 2 copies each of 10 related genes! Other proboscideans have fewer, but still more than the 2 copies of a single p53 gene that humans have.
Other long-lived or large-bodied organisms appear to fight cancer differently. For example, the naked mole rat (Figure 24) is extraordinarily long-lived for its body size, living up to three decades. The naked mole rat’s physiology blocks one of the hallmarks of cancer: metastasis.
In most organisms, healthy cells are controlled by contact inhibition: that is, when they come into contact with other cells, they stop dividing. Metastasizing cancer cells lose their contact inhibition. Even when grown in the lab, healthy cells stop dividing when they cover the surface of their dish, but cancer cells will keep dividing, piling on top of one another.
The cells of the naked mole rat, on the other hand, seem to be hypersensitive to contact inhibition.[1] They are also resistant to the epigenetic reprogramming that accompanies de-differentiation during cancer progression.[2]

Research on other large-bodied or long-lived species is likely to yield more insights into cancer physiology.
Test Your Understanding
Media Attributions
- Figure 23 DNA Repair © Sulak et al, 2016 is licensed under a CC BY (Attribution) license
- Figure 24 DNA Repair © By Roman Klementschitz, Wien is licensed under a CC BY-SA (Attribution ShareAlike) license

