Gene structure
The codons make up the coding sequence of a gene, but it’s important to remember that the coding sequence is not the only important part of the gene. All the regulatory elements we’ve discussed in the transcription and translation modules play a role in the function of the gene (Figure 18).
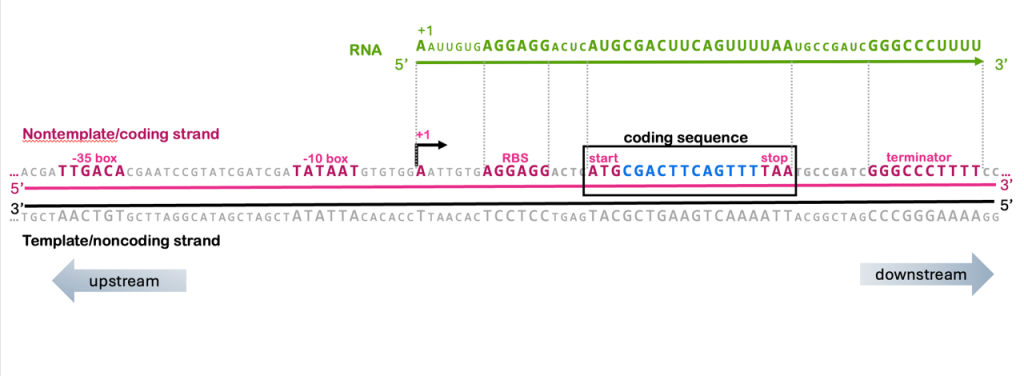
You’ll remember from the unit on transcription that the RNA molecule has the same sequence as the nontemplate strand of the gene. The nontemplate strand is also called the coding strand because it contains the coding sequence of the gene, and the template strand is also called the noncoding strand.
The translation control elements discussed in this module are also therefore found in the coding strand of the DNA: the ribosome binding site (AGGAGG), the start codon (ATG), and the stop codons (TAA, TAG, TGA) are all recognizable elements in the coding strand of a gene, as shown in Figure 18, you can even find codon tables that use DNA codons rather than RNA codons.
The coding sequence is also called an open reading frame (ORF) because it is a long stretch of codons that lack a reading frame. For simplicity, the ORF in Figure 18 is only 6 codons long, but most ORFs are hundreds or thousands of codons.
A note on terminology: What’s the difference between the ORF and the coding sequence? The term ORF is used when searching genomic DNA sequences for potential “new” genes. Because 3/64 codons are stop codons, if reading random, non-coding genomic sequences, you’d expect to find a codon about every 20 codons. If a long stretch of DNA – say, hundreds or thousands of codons – does not have a stop, that is called an “open” reading frame, and it is potentially part of a protein-coding gene. All coding sequences are ORFs, but not all ORFs end up being genes.
With this understanding of the relationship between coding strand, template strand, and RNA, we can therefore build a more complete understanding of the structure of a gene. In Figure 18, you can see that the RNA includes the sequence from the +1 to the end of the terminator, and all of the translation control elements (ribosome binding, start, and stop) are also within those boundaries.
Figure 19 shows the structure of a typical eukaryotic gene[1], reading from 5’ to 3’ on the coding strand. The transcribed region is shown between the two light blue regions, starting with the +1 site (labeled) and ending at the pA cleavage site (not labeled). The promoter sequences are shown upstream of the +1 site, with TATA box, CAAT box, and GC box drawn for this gene. The yellow boxes are enhancers, and you can see multiple enhancers upstream, downstream, and within the gene, as is typical for enhancers.
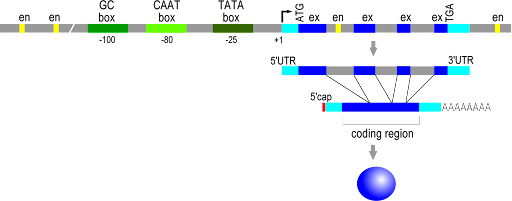
For both the prokaryotic and eukaryotic genes, notice that the control elements for translation are all within the boundaries of the transcribed region, that is, between the +1 site and the polyadenylated sequence, or between the +1 site and the terminator. This makes sense! A codon must be part of the RNA to be useful, so the start codon and stop codons must be between the +1 and the polyA site.
ORFs vs coding sequence revisited: In prokaryotes, the coding sequence is continuous, so there is one ORF for the gene. However, in eukaryotes, the coding sequence is discontinuous in the DNA sequence. The ORFs do not extend through the introns, from exon to exon, because the intron sequence will contain stop codons in frame with the coding sequence. Eukaryotic genes, therefore, include multiple ORFs.
Test Your Understanding
Media Attributions
- Diagram of a prokaryotic gene is licensed under a CC BY-SA (Attribution ShareAlike) license
- Structure of a typical eukaryotic gene © Open Genetics Lectures (OGL) Fall 2015 is licensed under a CC0 (Creative Commons Zero) license
- Locke, J. ‘Open Genetics Lectures’ textbook for an Introduction to Molecular Genetics and Heredity (BIOL 207). (2017) doi:10.7939/DVN/XMUPO6. ↵

