Haplotypes
It’s important to recognize that linkage studies like the HD RFLP analysis do not identify a causative gene, so the coinheritance of the chromosome 14 RFLP did not mean that polymorphism was within the HTT gene. Rather, the polymorphism was close enough to the gene that recombination did not occur in these families. Much more work was done to pinpoint the HTT more accurately within chromosome 4.
Like any DNA variant, polymorphisms arise due to a new (or de novo) mutation in a single individual. As that individual reproduces, the new variant may spread through the population. But remember that recombination is relatively rare between sites that are close together. So any new polymorphism tends to be inherited along with the sequence information that surrounds this. The new polymorphism would only be separated from surrounding polymorphisms due to recombination.
As a result, when we compare sequence information within a population of individuals, we tend to see blocks of sequence that are inherited together. For example, there are three polymorphic sites in the TAS2R38 gene, which encodes a bitter taste receptor. These SNPs affect the protein-coding region of the gene: position 145 of the gene specifies either a proline or alanine in the protein depending on the variant, position 785 specifies alanine or valine, and 886 valine or isoleucine. The alleles of the gene are named for the amino acid specified by each SNP. For example, PAV has a proline-alanine-valine combination, and AVI has an alanine-valine-isoleucine combination.
One might expect that, due to recombination, we would see all possible combinations of SNPs: PAV, PAI, PVI, etc. But, in fact, two are by far the most common, with about 95% of the human population having either PAV or AVI. People with the AVI variant usually cannot taste certain bitter substances, while people with the PAV variant can.
When we see certain morphs co-occurring more often than we’d expect, like the PAV or AVI combinations, we say that the loci are in linkage disequilibrium. The morphs that co-occur are said to be part of a haplotype. A haplotype is a group of SNPs (or other molecular markers) that tends to be inherited together in a block, as shown in Figure 21.
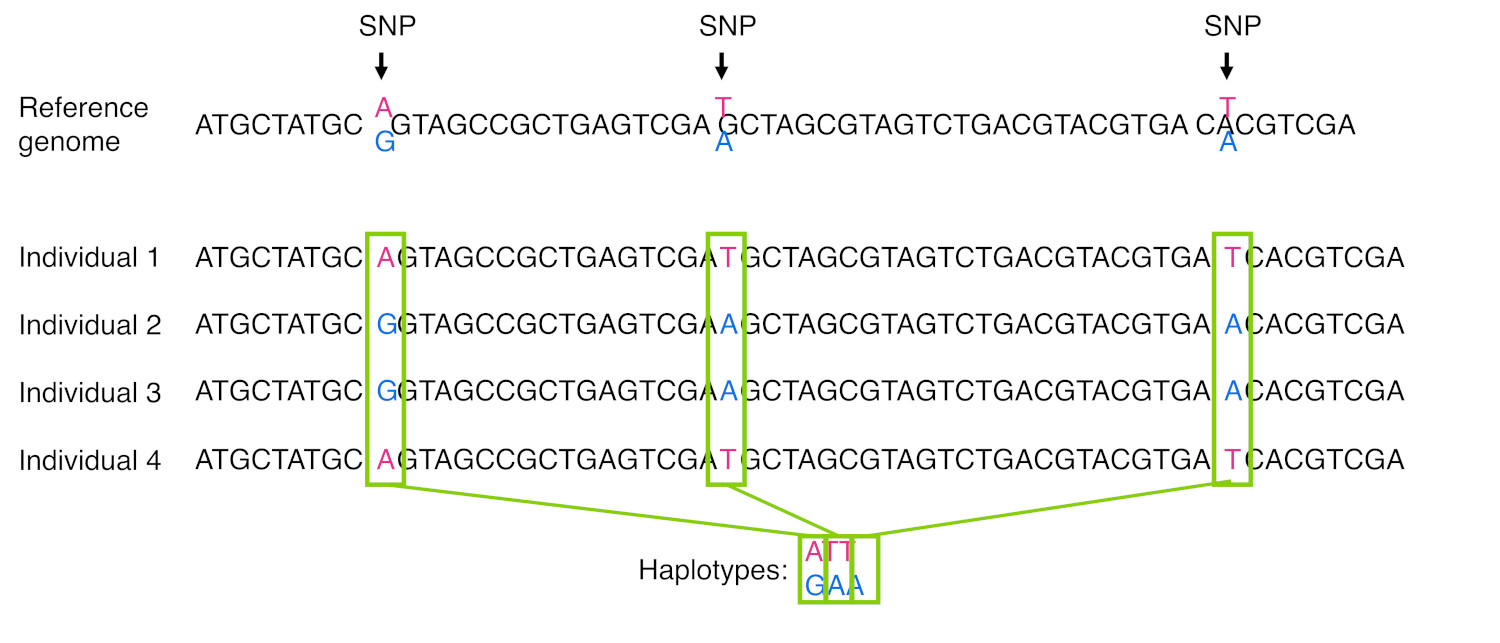
Haplotypes often arise when a relatively recent mutation event occurs and the variant becomes fixed in the population. Although recombination can separate that new mutation from surrounding SNPs, that may take many, many generations for that to occur. We tend to find that certain haplotypes occur more often in some populations than others, so haplotypes can be used to infer ancestry.
SNPs can be measured via a variety of molecular methods, including specialized versions of PCR and microarray analysis. There are about 5 million single nucleotide variants in the human genome. A SNP microarray or “SNP chip” may be able to measure an individual’s genotype at up to 2,000,000 SNPs with one test. Below is an animated GIF showing SNP microarrays.
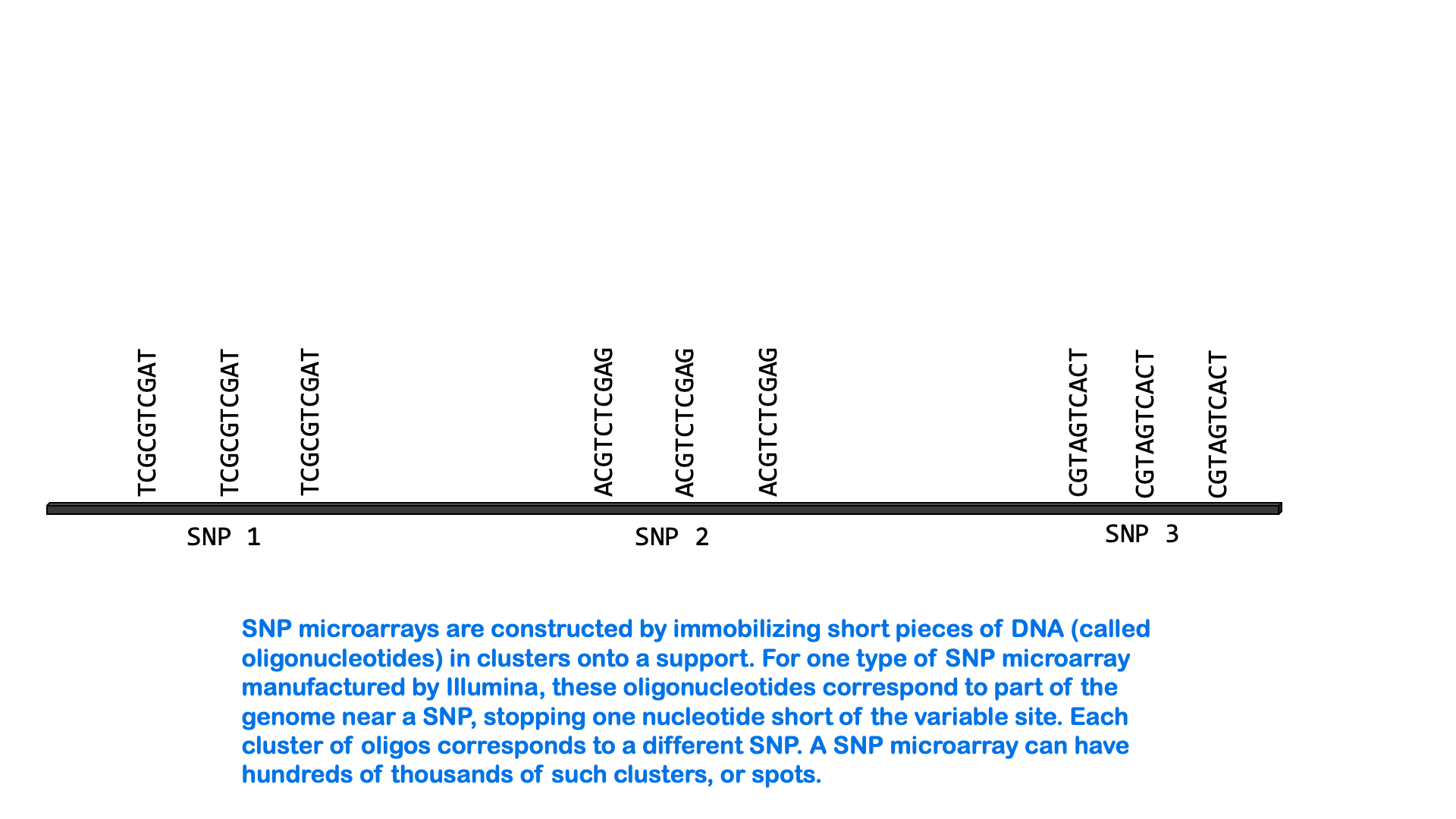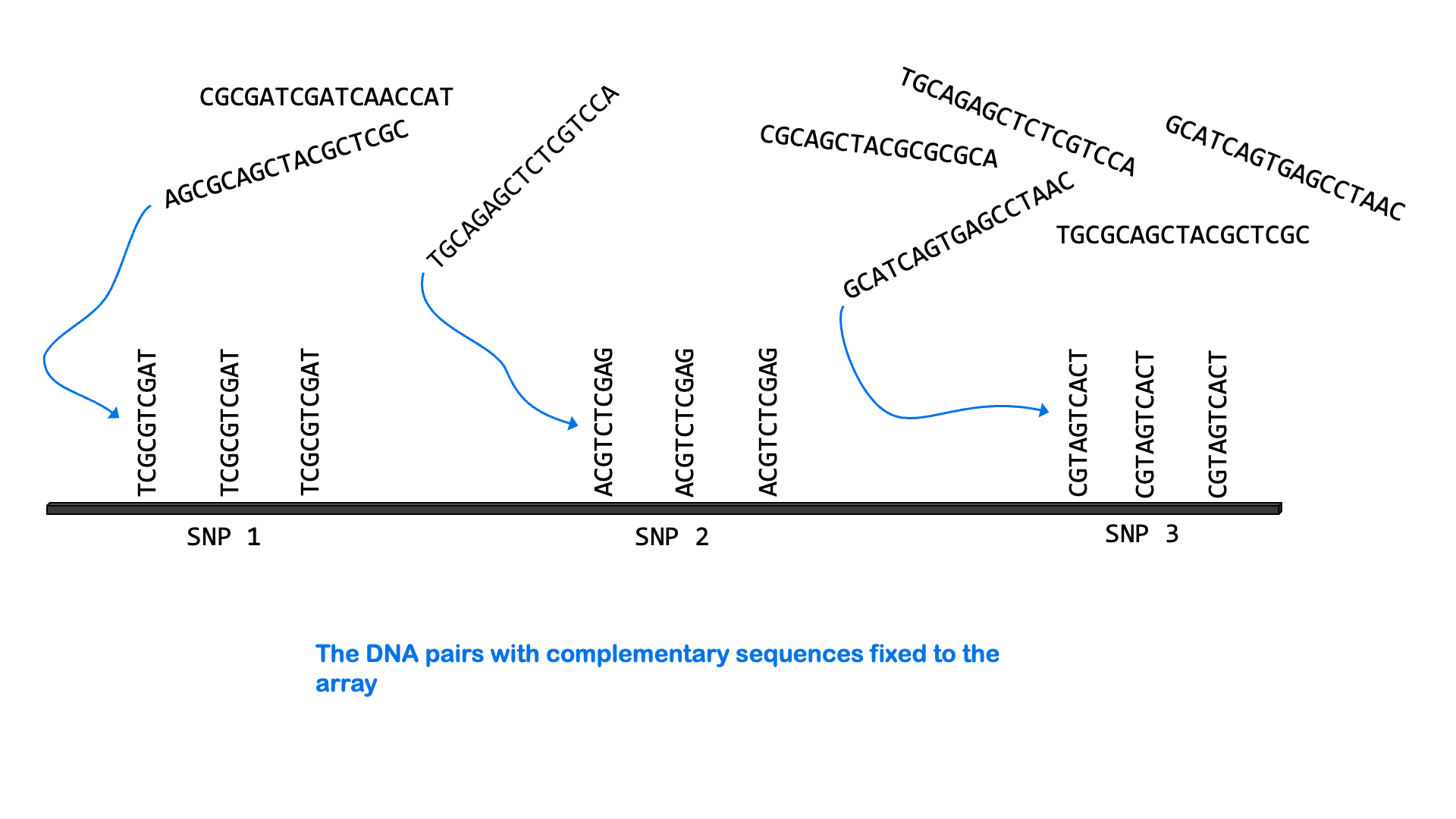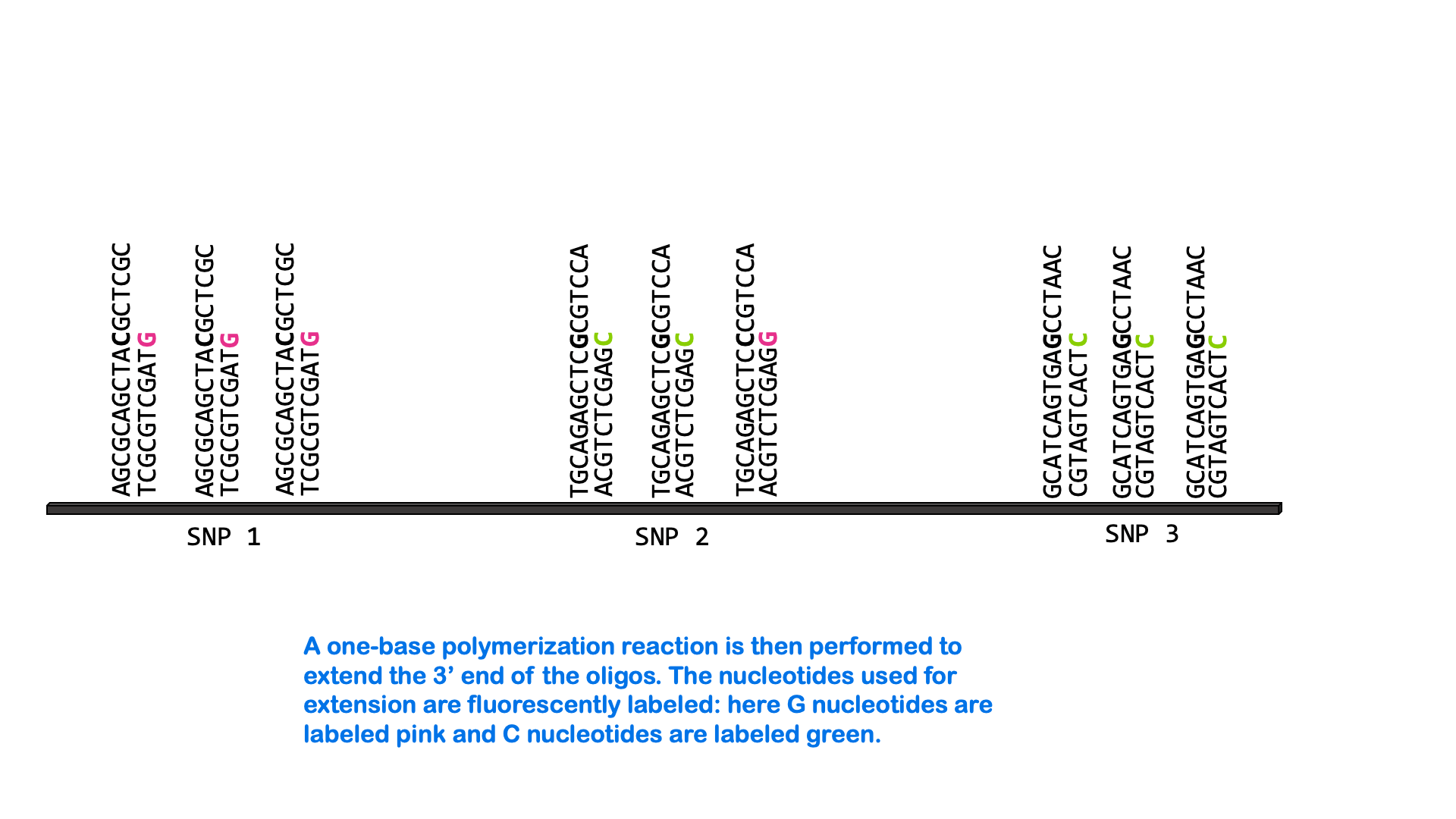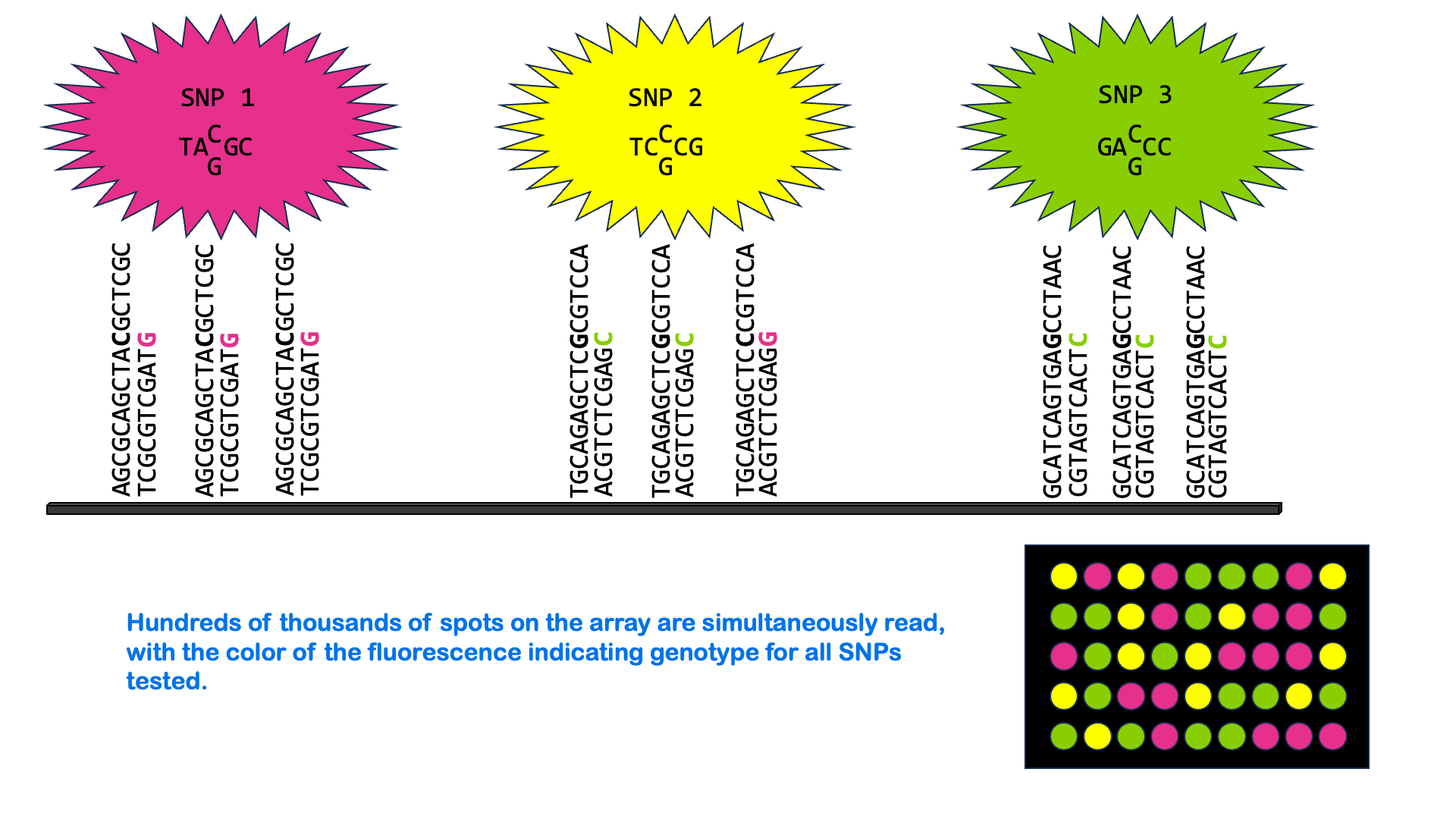
SNP arrays are used for genome-wide association studies (discussed more in the next section) as well as other purposes, including personalized medicine. Most of the commercially available direct-to-consumer DNA test kits use SNP microarrays to test for ancestry and health-associated genetic variants. But it’s not just individual SNPs that are used to determine ancestry. Very few SNPs belong to a single ancestral group, and no single SNP can be used to infer ancestry. Instead, these tests look for haplotypes and combinations of SNPs that are most common in people of certain ancestry.
Media Attributions
- Haplotypes © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license

