How do we know what enhancers do? Reporter genes experiments demonstrate promoter activity.
How do we know what the enhancer does? One way is to introduce a modified version of the enhancer sequence into the organism’s genome, fused to a reporter gene.
Reporter genes produce protein products easily visualized in the lab, often through a change in color. Some examples of reporter genes are Green Fluorescent Protein (which makes some jellyfish fluoresce green), luciferase (the enzyme responsible for fireflies “glowing”), and beta-galactosidase (which bacteria use to break down lactose, discussed with the lac operon in the Overview of Gene Regulation chapter). What’s important is that the expression of these genes is easy to detect, and these genes are not normally present in the cell or system we are studying so there’s no background signal to confuse.
When fused to a promoter or an enhancer we’re interested in studying, the reporter gene can “report” whether the sequence is transcribed and translated by the factors normally present in the cell.
For many of the eve experiments, a eukaryotic promoter with the stripe 2 enhancer DNA was fused with the bacterial LacZ gene sequence. The engineered DNA construct was then reintroduced into Drosophila embryos, and the expression of LacZ was monitored. Because the promoter controlling the LacZ gene was fused just with the stripe 2 enhancer, LacZ is expressed in the same region of the embryo as the eve stripe 2 but not in the other stripes. This type of experiment confirms the role of this segment of DNA in regulating eve expression in stripe 2. An example of these types of experiments is illustrated in Figure 12.
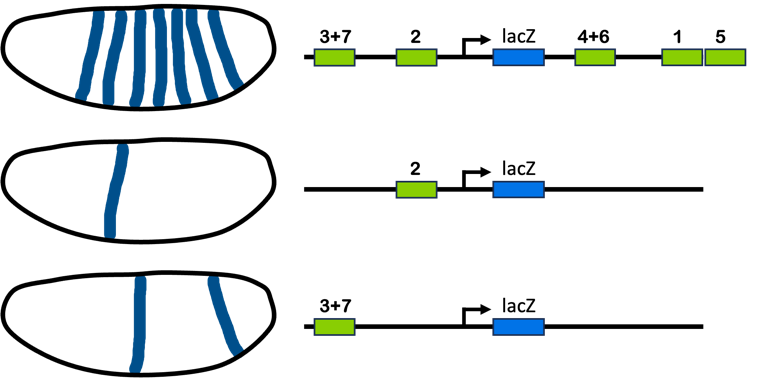
But it goes further than this! By engineering enhancers with parts missing or by crossing with flies that are mutant for other genes, we can break down the enhancer into its component parts and identify sequences important for function. For example, LacZ expression vanishes if the bicoid binding sites are mutated in the repressor. The width of the LacZ stripe expands toward the anterior pole if the enhancer is mutated to eliminate Gt binding sites if the reporter gene is expressed in an embryo that is mutant for giant . [1] [2] These kinds of experiments are examples of reverse genetics, where the genome of an organism is specifically manipulated to see an effect on phenotype.
Media Attributions
- Figure 12 EG Regulation © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license
- Arnosti, D. N., Barolo, S., Levine, M. & Small, S. The eve stripe 2 enhancer employs multiple modes of transcriptional synergy. Development 122, 205–214 (1996). ↵
- Goto, T., Macdonald, P. & Maniatis, T. Early and late periodic patterns of even-skipped expression are controlled by distinct regulatory elements that respond to different spatial cues. Cell 57, 413–422 (1989). ↵

