Human ancestry tests
We started this chapter with an example of genetic variation and adaptation to high altitude in humans. Let’s return to human ancestry, now with an understanding of the underlying mechanisms that affect evolution.
Humans, like all living things, can be better understood within the context of evolution.
The most ancient human populations likely arose in Africa, with several successive migrations from Africa populating other parts of the world. As these migrations occurred, there is evidence that the dispersing Homo sapiens interbred with Neanderthals and Denisovans , now-extinct hominin species that coexisted with early Homo sapiens.
The migrating populations continued to acquire genetic variants. Selective pressures in different geographic regions likely influenced which variants were maintained in different populations. For example, darker skin is usually found among people whose ancestors lived near the equator, and lighter skin is usually found among people whose ancestors lived in areas nearer to the poles. These adaptations confer protection against UV sun damage among darker-skinned populations near the equator, while lighter skin may be an adaptation that makes it easier for Northern populations to synthesize vitamin D.
The adaptation to high altitude described at the beginning of this chapter is another example. Genetic drift played a role in the accumulation of variants as well, with phenotypically neutral SNPs accumulating in some populations and not others due to the randomness of time and reproduction.
The migration of early humans was not unidirectionally “out” of Africa: there is evidence of “reverse migration” as well. Populations of early humans thus did not exist as discrete, reproductively isolated groups. Because of this, humans show considerable admixture among populations of different geographic ancestry. Rather than discrete ethnic groups, human variation appears to exist on a geographic continuum. Any one person is more likely to be similar to someone with ancestry from a nearby geographic location and less similar to someone of ancestry separated by more distance but rarely is there evidence of true reproductive isolation.
Human ancestry tests marketed by companies like 23andme, AncestryDNA, and others typically take advantage of the geographic association of certain variants; that is, the frequency of SNPs varies with geographic ancestry. These companies compare SNPs across the genome, looking for patterns that are similar to other groups of people of known ancestry.
However, single SNPs cannot predict ancestry. Instead, these genome analyses typically look at haplotypes, longer stretches of adjacent SNPs that tend to remain linked in multiple generations. (Haplotypes were first discussed in the chapter on linkage.) These ancestry tests also look at maternal lineage through mitochondrial DNA and paternal lineage through Y chromosome DNA. Because there is no recombination in mtDNA and the Y chromosome, these chromosomes are passed relatively intact from generation to generation. Closely related mtDNA and Y chromosome DNA arise from shared ancestry and are called haplogroups. A simple video overview of haplotypes and haplogroups can be found at Learn.Genetics.
Ancestry-testing companies look for the frequency of those haplotypes and haplogroups in reference populations of known ancestry. This frequency can predict the likelihood that someone of that geographic ancestry will have those combinations of SNPs. The larger the reference pool, the more reliable the test becomes. Because of this, such ancestry tests can pinpoint smaller and smaller geographic regions every year, but they are still limited by the people who have contributed to the reference pool. Historically, such companies have had more European reference data than for other continents, although this is gradually changing.
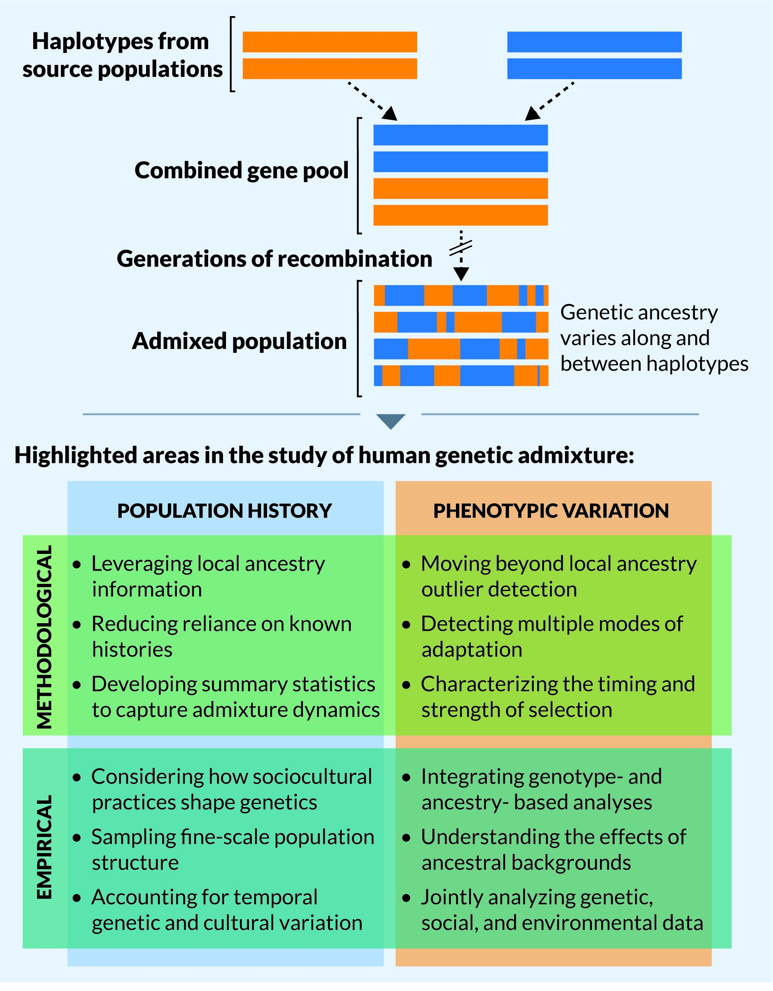
By looking at short stretches of haplotypes along a chromosome, these tests can also detect the admixture that results from ancestors of varying geographic ancestry, illustrated in Figure 16 (top).
Test Your Understanding
Media Attributions
- Figure 16 Haplotypes © Korunes and Goldberg, 2023

