Mechanism of replication
The basic chemical reaction of DNA synthesis is the addition of a nucleotide residue to the 3’ end of a polypeptide chain. The reaction is catalyzed by a DNA polymerase. But DNA polymerases have a biochemical limitations: they can only add nucleotides to the 3’ end of a strand that already exists. They cannot add nucleotides to the 5’ end of a molecule, and they cannot synthesize DNA de novo, or from the beginning by linking together two nucleotides. Therefore, to perform DNA synthesize with fidelity on both strands of the template DNA requires multiple polymerases and a collection of other proteins and enzymes. These include:
- DnaA: Recognizes the origin of replication, where the process begins
- Helicase: Unwinds the double helix, allowing the replication machinery to access the template DNA
- SSB: Holds the unwound strands of the double helix apart long enough for replication to occur
- Primase: Seeds the replication process, by synthesizing a short RNA molecule called a primer complementary to the template strand, since DNA polymerase can’t synthesize DNA de novo.
- DNA Polymerase III: The main replicative polymerase
- DNA Polymerase I: Removes the RNA primers and fills in DNA across the gap in the daughter strands
- Clamp: Keeps the polymerase from dissociating from the template DNA, allowing replication to continue for a long period of time without reloading the polymerase onto the DNA
- Ligase: Seals nicks in the phosphodiester bond that remain after the removal of RNA primers
Together, these proteins and enzymes make up the replisome, or replication machinery. Let’s look at each of them in turn. This next section is organized with a series of static figures that highlight what is happening to the DNA at each step of the process. The section concludes with a link to an animated GIF that shows the action of each of the proteins of the replisome.
Replication begins at specific sites in the DNA called origins of replication. Prokaryotes, which have a single circular chromosome, have a single chromosomal origin of replication. The origin includes a specific DNA sequence called oriC. In E. coli, this origin of replication is approximately 245 base pairs long and is rich in AT sequences. The oriC sequence is recognized by an initiator protein called DnaA, which binds to the origin in a sequence-specific manner. DnaA uses the energy of ATP hydrolysis to introduce torsional pressure that partially unwinds the double helix at the origin.
From the origin of replication, helicase enzymes extend the unwinding, again using the energy from the hydrolysis of ATP to break the hydrogen bonds connecting the strands of the double helix. Helicases move bidirectionally away from the origin, separating the two strands of the double helix into what is called a replication bubble. The Y-shaped edges of the bubble, where the DNA transitions from single-strand to double-strand, are called replication forks. Each replication bubble includes two replication forks (Figure 6). As replication proceeds, the two replication forks move continuously away from each other as the helicases continue to unwind more of the chromosome.
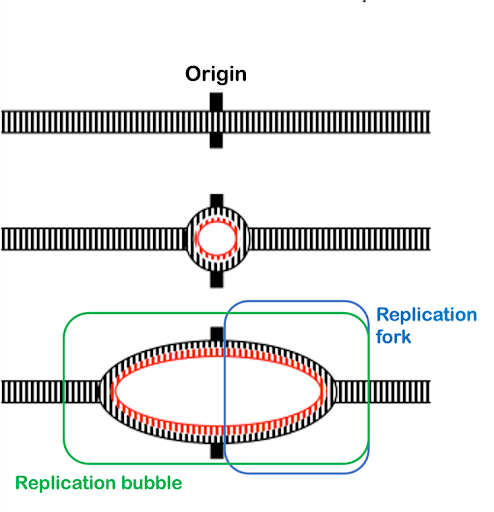
Re-pairing of the bases is energetically favorable and would happen spontaneously in an aqueous environment like a cell. But the replication machinery needs access to single-stranded DNA template. Single-stranded binding protein (SSB) coats the single-stranded DNA, holding the strands apart and preventing them from re-pairing.
Because DNA polymerases cannot synthesize DNA de novo, an additional enzyme is necessary to begin synthesis. Primase synthesizes short RNAs called primers complementary to the DNA near the origin. These short RNA molecules provide a free 3’ end for the DNA polymerase.
There are multiple DNA polymerases, but DNA polymerase III (Pol III) is the main replicative polymerase in prokaryotes. It performs most of the DNA synthesis at the replication fork. DNA pol III extends the primers outward from the origin toward the replication forks by adding nucleotides to the 3’ end, as shown in Figure 7.
The continuous synthesis from the 3’ end of the original primers form what are called the leading strands. Leading strand synthesis will continue for millions of bases! But Pol III doesn’t hold on to DNA very well: by itself, it would fall off within a handful of nucleotides. So a ring-shaped protein called the sliding clamp holds the polymerase into place on the template strand, making DNA synthesis highly efficient. The sliding clamp works almost like a carabiner, opening and closing to encircle the DNA. An enzyme called the clamp loader opens and closes the ring to engage clamp on the DNA. (Clamp loader is not shown in the images in this text.)
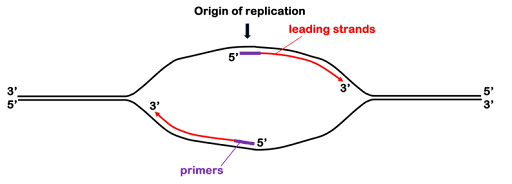
Prokaryotes can replicate about one thousand nucleotides of template DNA per second! Leading strand polymerization will continue until the polymerase runs out of unreplicated template, either because it bumps against another replication fork proceeding in the opposing direction or because it comes to the end of a linear chromosome (in eukaryotes).
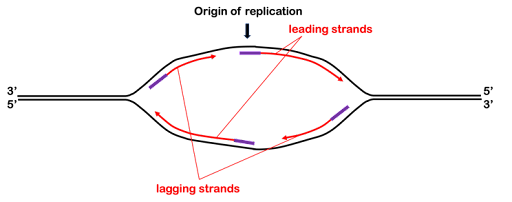
In Figure 7, you can see that the synthesis of the leading strands leaves half of the replication bubble unreplicated: DNA polymerase cannot extend the original primers on the 5’ end. Thus, an additional primer is placed on each strand nearer the replication fork. The polymerase synthesizes the lagging strand from this second primer, extending the 3’ end back toward the original RNA primer. This is shown in Figure 8. The 3’ ends of the daughter strands are indicated by arrowheads. You can see that the leading strand “points” toward the replication fork, while the lagging strand points away from the fork.
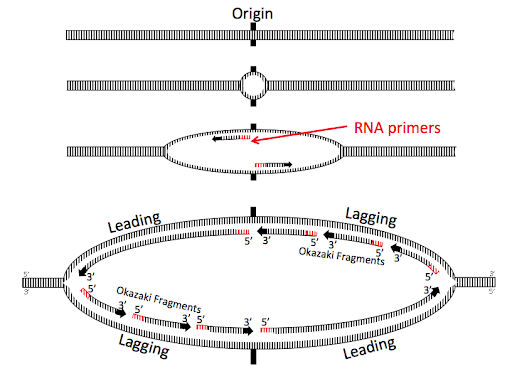
As the helicases continue to open the replication bubble, additional primers are created, and additional fragments of the lagging strand are synthesized. These lagging strand fragments are called Okazaki fragments after the married researchers who first hypothesized and confirmed their existence. Figure 9 shows a replication bubble with leading and lagging strands. All 5’ and 3’ ends are labeled, and the 3’ ends of the daughter DNA strands are indicated with arrowheads.
You’ll see the original RNA primers at the origin near the center of the replication bubble. These original primers form the 5’ end of two leading strands, synthesized continuously away from the origin.
The top right and bottom left of the replication bubbles are the lagging strands, with three Okazaki fragments shown for each lagging strand. The oldest Okazaki fragments are closest to the origin; the newest are closest to the forks.
Note the diagonal symmetry of the bubble: one leading strand is on the top left of the bubble, and the other is on the bottom right. The 3’ ends of all of the daughter strands on the top half of the bubble are oriented to the left, antiparallel to the 3’ end of the parent strand. The opposite is true for the bottom of the bubble. This geometry happens because of the antiparallel orientation of the strands of the double helix.
The lagging strand polymerase extends the 3’ end of each new RNA primer toward the previous Okazaki fragment, but the RNA is not retained in the genome. DNA polymerase I acts to remove the RNA and patch the space with DNA nucleotides. DNA polymerase I has a 5’ to 3’ exonuclease function that removes the RNA primer while it at the same time extends the 3’ end of the newest Okazaki fragment into the newly created gap. Fragments are linked together through the action of DNA ligase.
Test Your Understanding
Media Attributions
- Replication bubble © Online Open Genetics is licensed under a CC BY-NC-SA (Attribution NonCommercial ShareAlike) license
- The leading strands © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license
- The lagging strands © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license
- Origin of replication © Online Open Genetics is licensed under a CC BY-NC-SA (Attribution NonCommercial ShareAlike) license
- Stepped replication fork images © Amanda Simons is licensed under a CC BY-NC-SA (Attribution NonCommercial ShareAlike) license

