Methylation of DNA is often associated with decreased transcription

In addition to modification of histones, DNA itself can be epigenetically modified via methylation of cytosine. Cytosines in DNA can be methylated at position 5 of the base. The methylation does not change the underlying sequence of the DNA: Methylated cytosines still base pair with guanine, because the base-pairing parts of the molecule are unchanged. This is illustrated in Figure 7. But the methylation changes the part of the base that is facing the major groove, so methylation affects how transcription factors can interact with the major groove of DNA.
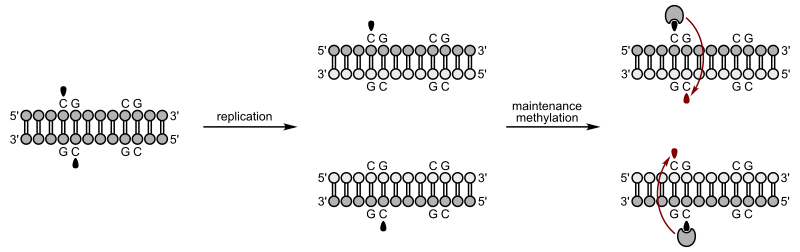
Methylation of cytosines typically occurs at specific positions within the genome. Potential methylation sites usually have a cytosine immediately followed by a guanine. These are called CpG sites, where the p represents the phosphate between the two sugars. This is a palindromic sequence, with the complementary strand also having the sequence CpG. Usually both strands are methylated, if a site is methylated at all, as shown in Figure 8. Regions of the genome with many clustered CpG sites known to function in gene regulation are sometimes called CpG islands.
Methylation of DNA itself is generally associated with decreased transcription regulation, including the silencing of highly methylated regions of the genome. The converse statement is also true: transcriptionally silenced regions of the genome tend to be highly methylated.
As with histone modification, DNA methylation is maintained after replication. Immediately after replication, the daughter double helix is methylated only on one strand. But DNA methyltransferases can recognize this hemimethylated DNA and modify the second strand to match. This is shown in Figure 8.
Enzymes called DNA methyl transferases catalyze the transfer of methyl groups from the methyl donor SAM — the same substrate that donates methyl groups to histones. DNA methylation is more stable than histone methylation: demethylation seems to be relatively rare. Methyl groups can be passively eliminated from DNA if daughter strands are not methylated following DNA replication. Methyl groups can also be eliminated from DNA via a series of enzymatic reactions that ultimately remove the methylated cytosine and replace it with an unmethylated cytosine.
Test Your Understanding
Media Attributions
- Cytosine and 5-methylcytosin © Wikipedia is licensed under a CC BY-SA (Attribution ShareAlike) license
- Maintenance methylation © Wikipedia is licensed under a CC BY-SA (Attribution ShareAlike) license

