Part I: Types of mutations
Geneticists use different terms to describe mutations, depending on context. In this section, we will look at ways to define mutations by the type of cell affected, the change made to DNA, and the effect on phenotype. If the mutation affects a gene, the mutation may also be described by the effect on gene function. If the mutation occurs within the protein-coding sequence of a gene, the mutation can be described by its effect on protein structure.
Type of Cell: Germline vs somatic mutations
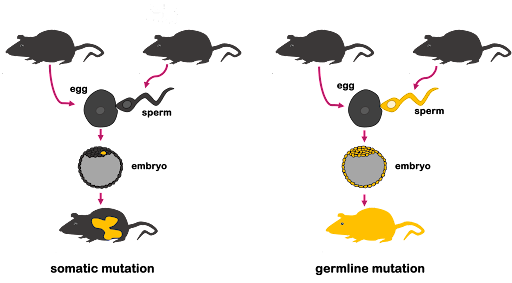
All mutations can be described by the type of cell that is mutated. Multicellular organisms that reproduce sexually use meiosis to produce haploid cells (egg and sperm) that come together to form a diploid zygote. That zygote undergoes mitosis, and successive cell divisions accumulate the cells that make up the embryo and, eventually, the full-grown organism.
If a cell in the embryo or full-grown organism sustains a mutation, that mutation can be passed along to all other cells that arise from mitosis of that cell. This is called a somatic mutation because it happens in somatic (nonreproductive) cells. A somatic mutation can result in just one or two cells that harbor the mutation, but if the mutant cell undergoes frequent mitosis, it might create a patchwork of mutant cells. This is shown on the left in Figure 2, where the mutation occurs in the embryo. A somatic mutation cannot be passed to offspring because the germ cells are not mutated.
If a mutation occurs in a germ cell (egg or sperm), the mutation will become part of the genome in the zygote. As the zygote undergoes mitosis, the mutation will be passed along to all daughter cells. Every cell in the mature organism will have the mutation, including its germ cells. So the mutation is called a germline mutation, and the mutation can be passed to the next generation of offspring. This is shown on the right in Figure 2. Germline mutations can also arise if a mutation occurs in the egg, the zygote, or in an early enough stage of the embryo that the reproductive cells of the organism contain the mutation.
Test Your Understanding
Changes to DNA: Insertions, deletions, base substitutions, and large-scale rearrangements
All mutations can be described by what happens to the DNA: additional bases can be added (an insertion), removed (a deletion), or one or more bases can be changed for different bases (a base substitution). These are illustrated in Figure 3.
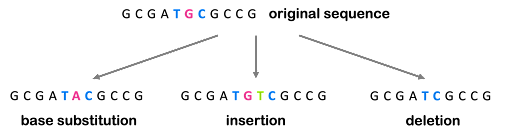
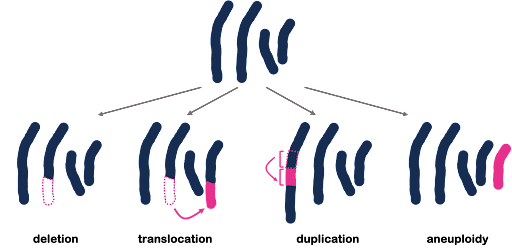
All three of the examples shown in Figure 3 are described as point mutations because they affect one point within the genome. But we also see mutations that affect a larger portion of the genome, called structural variants. These larger-scale chromosomal rearrangements can include the gain or loss of entire chromosomes, which is a change in chromosome number called aneuploidy. On the other hand, part of a chromosome can be lost, part of a chromosome can be duplicated, or part of a chromosome can be translocated to another. Examples are illustrated in Figure 4. Although not shown in Figure 4, inversions are segments of the chromosome that have been reversed or flipped in orientation within the rest of the sequence.
Changes to protein sequence: Frameshift, missense, nonsense, and silent mutations
Most mutations are not found within the protein-coding sequence. After all, in humans, only about 1% of the genome is protein-coding sequence! But if a point mutation occurs in the coding sequence for a protein, we can also describe the mutation with respect to how it changes protein sequence.
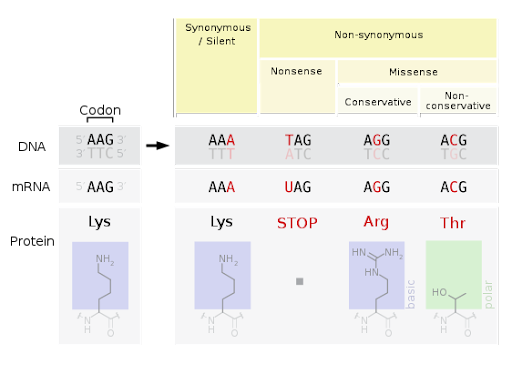
Base substitution mutations fall into one of three categories, illustrated in Figure 5.
- A silent mutation changes the DNA sequence but does not change the protein sequence. This is possible because most amino acids are specified by more than one codon. It is also called a synonymous mutation.
- A nonsense mutation changes an amino acid-specifying codon to a stop codon. This creates a protein that is shorter than normal because translation can’t continue past the stop codon.
- A missense mutation changes an amino acid for a different amino acid. Missense mutations can be conservative (changing an amino acid for one that is chemically similar) or nonconservative (like exchanging an acidic base for a basic base or a polar base for a nonpolar one).
Test Your Understanding
Insertion or deletion mutations may simply add or subtract amino acids from the protein, but only if the insertion is a multiple of three nucleotides. If the number of inserted or deleted nucleotides is not divisible by three, this will result in a frameshift. Remember that since the coding sequence is translated in codons of three bases, each DNA strand has three potential reading frames. Inserting or deleting one, two, four, or any number of bases that are not a multiple of three will cause the ribosome to translate the wrong frame downstream of the mutation. Every amino acid after the insertion or deletion point will be different. This is shown in Figure 6.
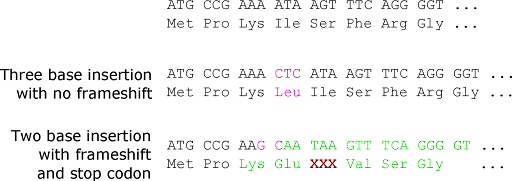
Test Your Understanding
Changes to gene function: Gain of function vs loss of function mutations
The terms silent, nonsense, missense, and frameshift only apply to mutations in the coding sequence of a gene. But mutations in DNA can occur anywhere in the genome – they can occur within genes, they can occur between genes, they can occur in coding sequences or in regulatory elements or in other noncoding regions. In fact, most mutations occur in non-coding sequences simply because the coding sequence is such a very small fraction of the genome. As a result, most mutations don’t have much effect – we call them neutral mutations.
Even mutations within a coding sequence can be neutral: Silent mutations and conservative missense mutations are often neutral mutations.
But sometimes mutations can affect the function of a gene – either because the coding sequence is altered or because an important regulatory element is affected. For example, a mutation might delete the promoter or change an intron sequence in a way that prevents proper splicing. In all of those cases, we describe the mutation in terms of how the mutation affects the function of a gene.
Test Your Understanding
A gain of function mutation is a mutation that causes a gene to do something extra. For example, in Figure 4, duplications that result in an extra copy of a gene are often gain of function mutations because an extra copy of a gene means more protein is produced. A mutation that prevents transcription from being turned off would be another example of a gain of function mutation. Gain of function mutations can also affect protein sequence: for example, if an inhibitory domain of a protein is altered, the whole protein could be active even if it is meant to be inactive. We see examples of this in the Gene Expression chapters of this text.
You might think that doing something extra is good! And some gain of function mutations may be beneficial to an organism. But not necessarily. Think of an analogy to a gas stove (Figure 7). If one of the burners makes an extra big flame and cannot be turned off, that could start a kitchen fire! As an example in human biology: gain of function mutations in certain proteins that trigger cell division are associated with cancer because the mutation causes cells to divide uncontrollably. (The cancer-causing gain of function mutations are somatic mutations, not germline.)

A loss of function mutation is a mutation that lessens the activity of a gene. This could be a complete loss of function – say if no RNA or protein is produced. If no functional protein is produced, it is also called a null mutation – “null” is a word that simply means “zero”. The chromosomal deletion in Figure 4 would cause a null mutation of all deleted genes, as would any mutation that destroyed the function of a gene’s promoter. To go back to the stove analogy in Figure 7, this would be like a burner that can’t turn on.
Some loss of function mutations reduce but don’t completely block the action of the gene. These partial loss of function mutations can still affect the phenotype of an organism.
For example, in humans, the disease cystic fibrosis is caused by a loss of function in the gene CFTR, which is important for Cl– transport across the cell membrane. Without functional CFTR protein, salt and water balance is impacted, and as a result, people with cystic fibrosis produce thick, sticky mucous that causes problems in the lungs and digestive tract.
Interestingly, some disease-associated CFTR mutations are not null: protein is produced, although it does not function. Medical researchers have developed drugs that restore some function to the altered protein, relieving CFTR symptoms. The first of these drugs, trade name Kalydeco (ivacaftor), was released in 2012. These drugs have improved the quality of life and dramatically increased life expectancy for people with that mutation. However, they only work for patients with a subset of CFTR mutations. These drugs do not work for patients with null mutations, for example.
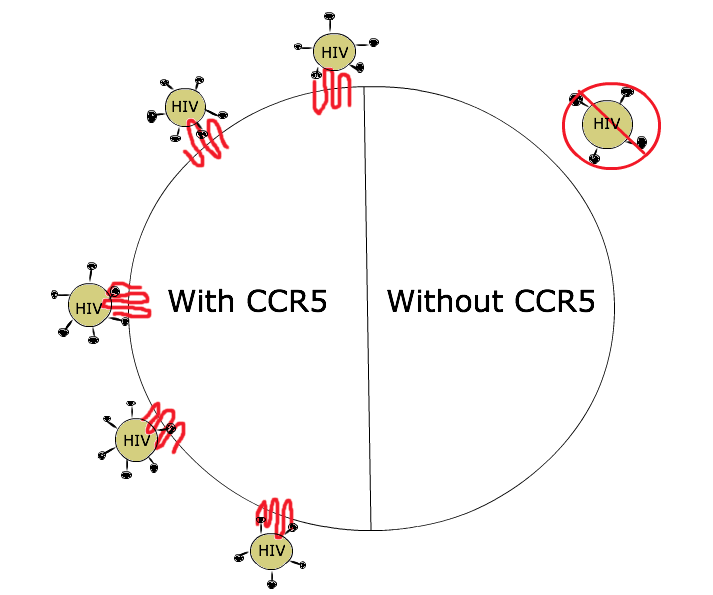
Loss of function does not imply that it’s good or bad! Loss of function mutations can be beneficial to an organism, too. For example: loss of function mutations in the CCR5 gene make people resistant to HIV infection. The normal CCR5 protein acts as a co-receptor, or attachment site, for the HIV virus to infect a cell, as shown in Figure 8. Without CCR5 protein in the membrane, there’s no infection. The mutations that confer resistance to HIV are found in the promoter (reducing transcription of the gene) and in the coding sequence of the gene (nonsense, missense, and frameshift mutations).
Test Your Understanding
In diploid organisms, gain of function mutations are usually, but not always, dominant: only one mutant allele is necessary to cause a phenotype. To use the stove analogy again: if one burner is on fire, it doesn’t matter what the backup burner is doing; the kitchen is still on fire.
Loss of function mutations are usually, but not always, recessive. Two mutant alleles are needed to produce the mutant phenotype. Back to the stoves again: if one burner isn’t working, you can still cook dinner on the other. But if both burners are out, no dinner is heated.
An exception to this dominant/recessive rule is seen in haploinsufficient genes. The word haploinsufficient means “half is insufficient” or “half is not enough.” These are genes for which one copy is not enough to produce a normal phenotype, usually because the quantity of protein is important. Note that in this case, the word haploinsufficient applies to the gene, not the mutation!
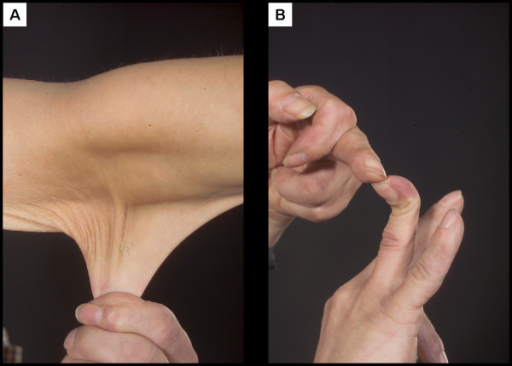
An example of a human trait caused by haploinsufficiency is Ehlers-Danlos Syndrome (EDS). People with EDS have overly loose joints prone to dislocation along with stretchy, elastic skin, which you can see in Figure 9[1]. EDS can be caused by a mutation in one of several genes, but the classic form of the syndrome is caused by a loss of function in the COL5A1 gene. This gene encodes a form of collagen, a fibrous protein important in connective tissue. With one less functional copy of the gene, less collagen is produced, leading to the phenotype[2].
Mutations in both coding and non-coding regions can affect the function of a protein. For example, if the sequence of the promoter is changed or deleted so the transcription machinery can’t bind to the promoter, no RNA or protein will be produced. This is a loss of function. If the sequence of the promoter is changed to increase the binding of the transcription machinery, the rate of transcription may be increased, leading to extra protein production. This is a gain of function.
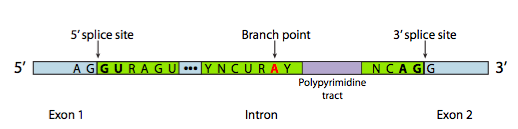
Even intron sequences can affect phenotype: the module on Transcription and RNA Processing explains how the removal of introns depends on consensus sequences at the intron/exon boundaries plus a “branch point site”, shown below in Figure 10. If those sequences are altered, the intron won’t be removed from the RNA. If the mRNA retains introns, it can’t be properly translated.
Test Your Understanding
Does the mutation change the effect of other mutations? Revertant and suppressor mutations
The examples so far have focused on single mutations. But in some cases, mutations can even undo the effects of other mutations! Such mutations are called suppressor mutations because they suppress, or block, the first mutation.
Suppressor mutations can be intragenic, undoing the effect of another mutation in the same gene, or intergenic, undoing the effect of a mutation in another gene.
Although there are many mechanisms by which suppressor mutations can act, one example is through “matching” mutations in molecular binding partners. For example, if two proteins interact to form a larger complex, as shown in Figure 11, a change to one protein might disrupt binding with its partner. But if a second mutation changes the partner in exactly the right way, this could allow binding to occur again.
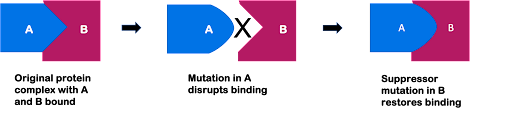
An example of this are the so-called nonsense suppressors. As discussed earlier in this module, a nonsense mutation changes an amino acid-specifying codon to a stop codon, resulting in a truncated (short) protein. A nonsense suppressor allows the translation machinery to read through a stop codon and continue synthesizing the rest of the protein! How does this work? Remember, the stop codon usually signals translational termination because it can’t be recognized by a tRNA. Nonsense suppressors are mutations in a gene encoding a tRNA! The tRNA sequence is mutated so that the anticodon is complementary to the stop codon.
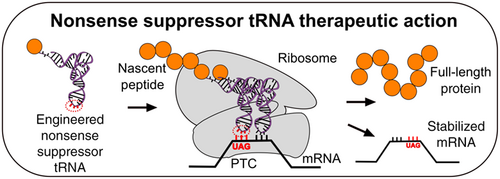
Although nonsense suppressors occur naturally, engineered nonsense suppressors are an interesting possibility for treating genetic diseases caused by nonsense mutations (Figure 12). Because many diseases are caused by nonsense mutations, it is hoped that a treatment using nonsense suppressor tRNA could be used to treat multiple diseases[3]. As of 2023, several companies are working to develop tRNA-based therapies for diseases caused by nonsense suppression[4].
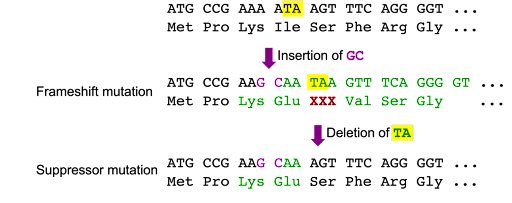
Intragenic suppressor mutations are a second mutation that undoes the effects of a first mutation. An example of this is shown in Figure 13, where an insertion of two bases causes a frameshift, but an insertion of two bases and a deletion of two bases results in a short change to the sequence but no frameshift.
The type of change to a protein determines the impact to protein function, as well as the location of the mutation. For example, a silent mutation is predicted to have no impact on the function of a protein. A conservative missense mutation is likely to have far less impact on the function of a protein than a frameshift, since only one codon is affected rather than many. And a frameshift early in a gene would likely to have greater impact to the function of the protein than would a frameshift that happens at the end of a gene, simply because more codons are affected.
Test Your Understanding
Media Attributions
- Germline vs somatic mutation © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license
- Mutations and DNA © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license
- Large-scale rearrangements © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license
- Base substitution mutations © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license
- Frameshift-mutation © L. Canham is licensed under a CC BY-NC (Attribution NonCommercial) license
- Two-gas-burners © By BogTar201213-Own work is licensed under a CC BY-SA (Attribution ShareAlike) license
- CCR5-vs.-CCR5-Mutation © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license
- Prominant-hyperextensible-skin-hypermobile-joints © Whitaker JK, Alexander P, Chau DY, Tint NL is licensed under a CC BY (Attribution) license
- Consensus-sequences-for-splicing © LibreTexts is licensed under a CC0 (Creative Commons Zero) license
- Protein-A-disrupts-binding- © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license
- Nonsense-suppressors © Porter et al 2021 is licensed under a CC BY (Attribution) license
- Intragenic-suppressor © L. Canham/modified by Amanda Simons is licensed under a CC BY-NC (Attribution NonCommercial) license
- Whitaker, J. K., Alexander, P., Chau, D. Y. & Tint, N. L. Severe conjunctivochalasis in association with classic type Ehlers-Danlos syndrome. BMC Ophthalmol. 12, 47 (2012). ↵
- Online Mendelian Inheritance in Man, OMIM®. Johns Hopkins University, Baltimore, MD. MIM Number: *120215 COLLAGEN, TYPE V, ALPHA-1; COL5A1. last edit 05/18/2021. World Wide Web URL: https://omim.org/. ↵
- Porter, J. J., Heil, C. S. & Lueck, J. D. Therapeutic promise of engineered nonsense suppressor tRNAs. WIREs RNA 12, e1641 (2021). ↵
- Elie, D. tRNA therapeutics burst onto startup scene. Nat. Biotechnol. 40, 283–286 (2022). ↵

