Prokaryotic genome structure
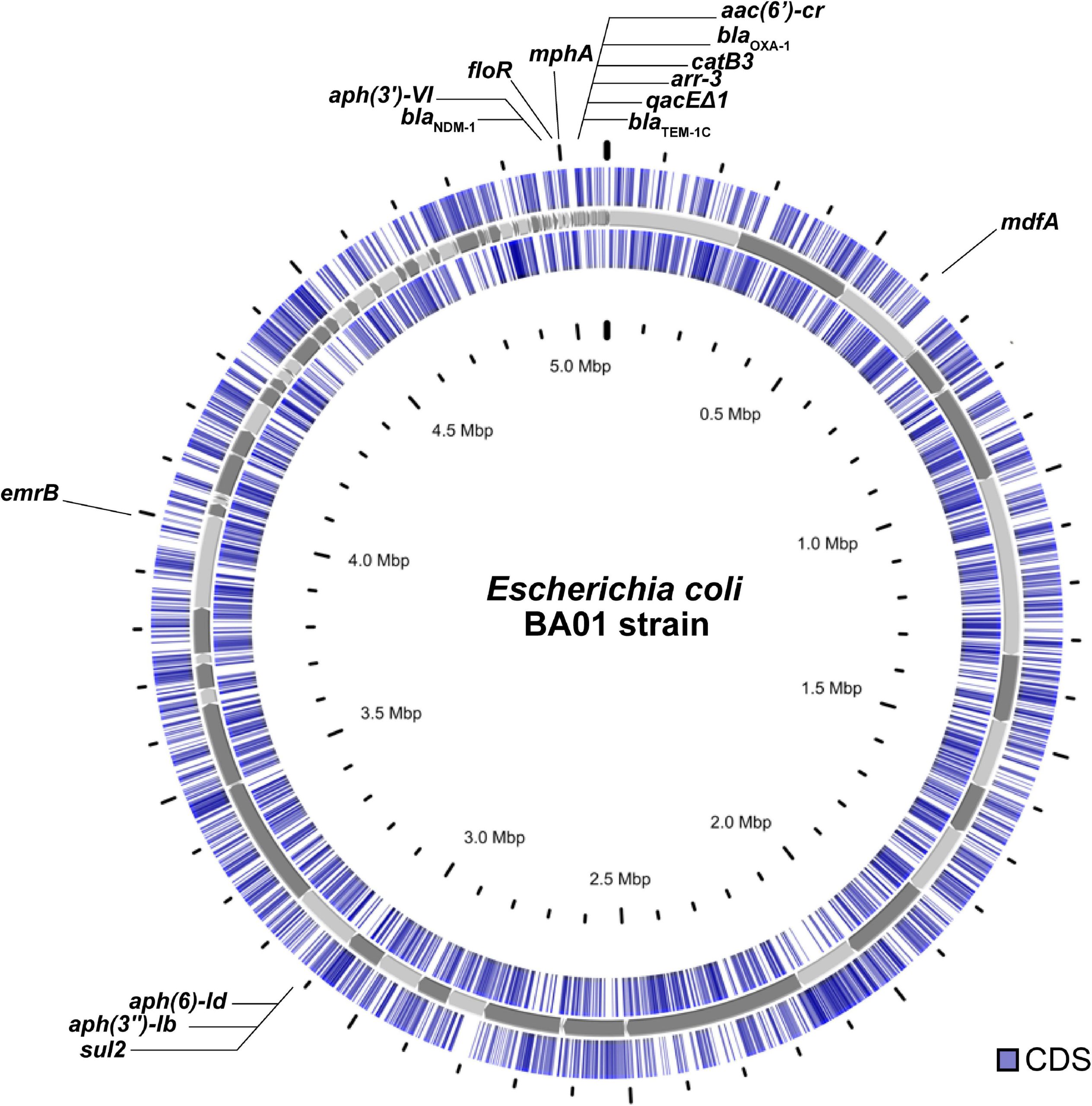
Figure 1 shows a diagram of the circular chromosome from one strain of the bacteria Escherichia coli, modified from a recent research paper. The E. coli genome is 5 million base pairs long, or 5 megabase pairs (5Mbp). There are about 5,000 genes within that chromosome. The diagram shows the location of the genes arranged along the chromosome, with the coding sequence of each gene, or cds for short, highlighted in blue. The coding sequence is the information that will be used to produce a protein (discussed more in later chapters). Remember that DNA is double-stranded: gene coding sequences can be on either strand, and the two concentric circles of blue marks indicate on which strand the coding sequence is found[1].
Bacterial genomes typically do not have very much DNA sequence separating genes. Depending on species, 90-95% of the genome may be coding sequence[2]. Some of the interspersed DNA includes regulatory sequence that is important for determining under which conditions a gene might be used to produce a protein.
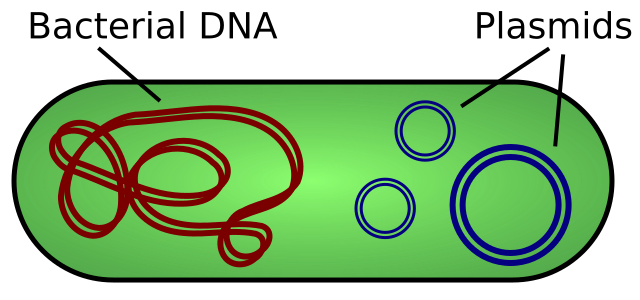
In addition to the circular genome, bacteria may also harbor additional extra-chromosomal DNA molecules called plasmids, illustrated in Figure 2. Each bacterial cell typically only houses one copy of the genome, but plasmids are usually present in multiple copies. Plasmid DNA, like the bacterial chromosome, is circular, but usually much smaller (measured in a few kilobases rather than megabases). Like the bacterial genome, plasmid DNA is copied by the cell and passed to daughter cells during cell division. Plasmid DNA can carry genes.
Antibiotic resistance genes are often encoded on plasmids, and the exchange of plasmids among bacterial cells can play a role in the spread of bacteria resistant to antibiotics. You can read more about the role plasmids play in the spread of antibiotic resistance in this article from the American Society for Microbiology[3].
Although not discussed extensively in this text, bacteria can, under some conditions, take up plasmid DNA from their environment or from another bacterial cell through a process called conjugation.
Plasmid DNA can also readily be transferred to both prokaryotic and eukaryotic cells in the lab, so plasmids are commonly used to study mechanisms of genetics.
Media Attributions
- Escherichia coli BA01 strain © frontiers is licensed under a CC BY-SA (Attribution ShareAlike) license
- Plasmid © User:Spaully via. Wikipedia is licensed under a CC BY-SA (Attribution ShareAlike) license
- Sellera, F. P. et al. Genomic Analysis of a Highly Virulent NDM-1-Producing Escherichia coli ST162 Infecting a Pygmy Sperm Whale (Kogia breviceps) in South America. Front. Microbiol. 13, (2022). ↵
- Rogozin, I. B. et al. Congruent evolution of different classes of non-coding DNA in prokaryotic genomes. Nucleic Acids Res 30, 4264–4271 (2002). ↵
- Plasmids and the Spread of Antibiotic Resistance Genes. ASM.org https://asm.org:443/Articles/2023/January/Plasmids-and-the-Spread-of-Antibiotic-Resistance-G. ↵

