Prokaryotic vs eukaryotic replication
Replication in eukaryotes uses the same chemistry of 5’ to 3’ synthesis as in prokaryotes, but there are notable differences that stem from the differences in prokaryotic and eukaryotic genome structure.
First, bacterial chromosomes are circular and relatively small compared to linear eukaryotic chromosomes. For example, the E coli genome is about 4,600,000 base pairs, or 4.6 megabases. Most bacterial chromosomes have a single origin of replication, and replication proceeds bidirectionally around the chromosome until the two replication forks meet. At 1000 basepairs per second, the whole bacterial chromosome can be replicated in a little more than half an hour.
In comparison, eukaryotic replication forks are much slower, moving at about 50 bases per second. The slower speed is at least in part because of the structure of the chromatin packaging. And eukaryotic chromosomes are much larger. Human chromosomes, for example, range from 48 megabases (Chromosome 21) to 249 megabases (Chromosome 1). If each eukaryotic chromosome were replicated only from one origin, at 50 base pairs per second, it would take about 5.5 days to replicate the smallest chromosome and about a month to replicate the longest one!
But eukaryotic cells don’t take that long to replicate a chromosome. Instead, eukaryotic chromosomes typically activate multiple origins along the length of each chromosomes, as shown in Figure 13.
Eukaryotic cells also use different enzymes, although they perform functions like those in prokaryotes. For example, while DNA pol III is the primary DNA polymerase in prokaryotes, in eukaryotes Pol epsilon and Pol delta perform the majority of leading strand and lagging synthesis, respectively. The sliding clamp function is performed by the eukaryotic protein PCNA. And rather than Pol I to replace the RNA primer with DNA, in eukaryotic systems RNAseH breaks down the RNA primers as Pol epsilon and delta continue their synthesis across the gap.
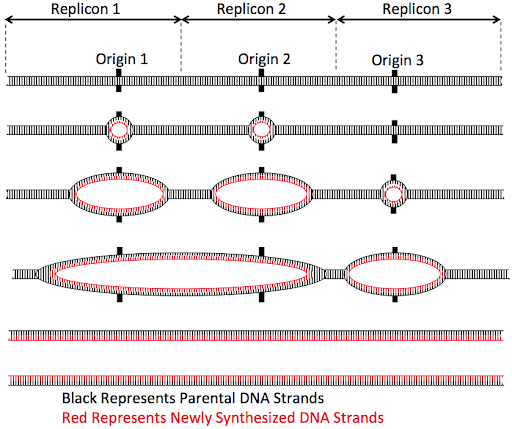
But perhaps the biggest difference between eukaryotic and prokaryotic replication comes from the fact that eukaryotic chromosomes are linear rather than circular. Because DNA replication requires a primer to begin, and because synthesis can only proceed in a 5’ to 3’ direction, eukaryotes have an “end replication problem”: they cannot replicate the 5’ most end of a linear chromosome. The 5’-most end of a newly synthesized daughter strand must always begin with an RNA primer. But if the 5’ RNA primer were degraded, there is no upstream 3’ end to extend. This leaves a gap at the 5’ end of the daughter strand, as shown in the top panel of Figure 14.
For most eukaryotic cells, this means that in every round of cell division, the ends of chromosomes, called telomeres, will get a little bit shorter. Older individuals typically have shorter telomeres than younger individuals of the same species. This shortening of telomeres is thought to play a role in cellular aging.
Telomeres are comprised of a repeating DNA sequence. In humans, the sequence is TTAGGG, but other species have slightly different sequences. The telomeric sequence can be repeated 100-1000 times at the ends of chromosomes. The telomeric sequences thus serve as a “cap” of sorts to the linear chromosomes: There are no genes in telomeric regions of the chromosome, so the DNA lost from the ends is not needed for gene expression.
However, some specialized cells – germ cells and stem cells – have an enzyme called telomerase that counteracts this problem. It is tempting to hypothesize that telomerase just works to fill in the 5’ gap. However, what telomerase actually does is make the template strand – already longer than the daughter strand – even longer!
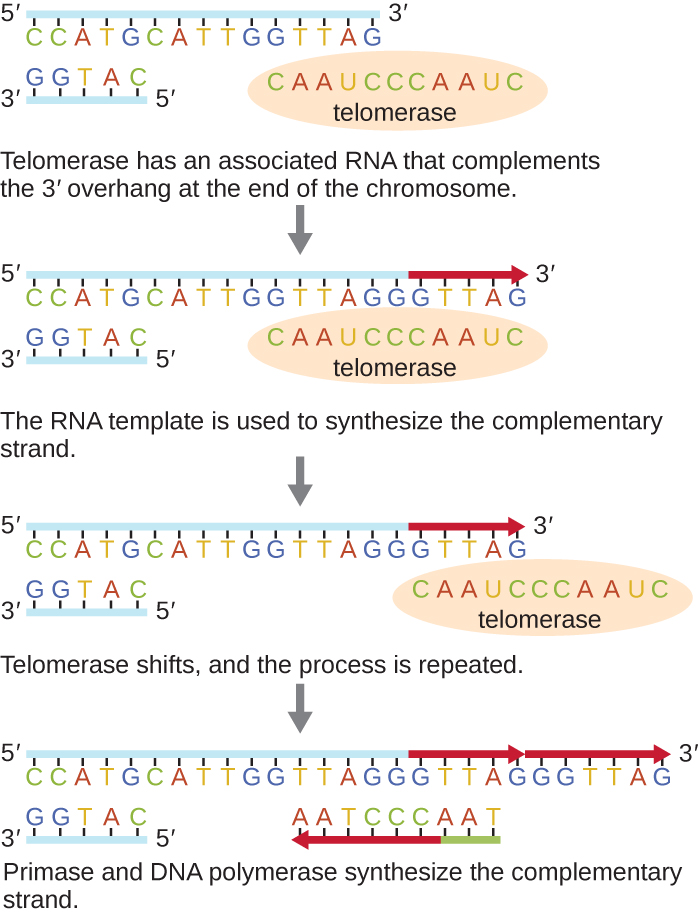
Telomerase does this by carrying its own template for DNA replication: an RNA molecule that is a component of the telomerase enzyme. The RNA molecule is complementary to the telomeric sequence. Telomerase binds to the 3’ end of the parent strand, with the RNA component base-pairing with the telomeric sequence in such a way that the telomerase RNA overlaps past the end (Figure 14). It supplies its own template for replication! The protein component of telomerase then adds additional nucleotides to the 3’ end of the parent strand, making it even longer. Telomerase then shifts toward the new 3’ end, and the process is repeated.
The additional length on the parent strand permits additional lagging strand fragments to be synthesized, thus lengthening the daughter strand as well. Note, however, that the daughter strand will still be shorter than the parent strand since that additional fragment also required a primer to initiate synthesis.
Test Your Understanding
Why do we care about telomerase? Telomerase, aging, and cancer
The 2009 Nobel Prize in Physiology or Medicine was awarded jointly to Elizabeth Blackburn, Carol Greider , and Jack Szostak for their work in understanding the mechanism of telomerase[1]. This reflects the profound implications of telomeres and telomerase for human medicine.
Telomerase may seem like an interesting trivia tidbit: after all, it’s only active in a very, very limited number of cells in an adult body. But telomerase is very important in “resetting” the genome to a youthful, long-telomeric state during gametogenesis – the production of egg and sperm for reproduction. Without the action of telomerase, each generation of offspring would have shorter and shorter telomeres. Telomerase is also active in stem cells. Stem cells have the potential to develop into other cell types, and they are self-renewing. Stem cells play an important role in replenishing tissues like blood and immune cells, as well as repairing tissues damaged by injury.
Mutations in the genes that encode telomerase (both the enzymatic component and the structural RNA component) are associated with phenotypes of premature aging. In humans, people who inherit rare mutations in these genes may show premature gray hair and other signs of aging, fibrosis of the lung and liver, immune dysfunction, and bone marrow failure. Other studies show that people with shorter telomeres than their same-aged peers are at higher risk of dying from certain causes, including cardiovascular, respiratory, and digestive diseases[2].
Because of these observations, medical researchers who study aging have hypothesized that telomerase-activating drugs might be useful in combating aging and age-related diseases. But it’s not so straightforward. One more type of cell often expresses high levels of telomerase: cancer cells.
Cancer cells cause disease because they divide uncontrollably, outgrowing their surrounding cells and destroying the function of healthy tissue. They also have the potential to metastasize, migrating throughout the body and colonizing secondary tumors in other tissues. Because telomeres get shorter with every round of cell division, most healthy cells can undergo a finite number of cell divisions before they senesce, or stop dividing. But many cancers express telomerase, allowing them to bypass this limit, continue to divide, and out-compete healthy cells in the body. This means that an anti-aging drug that activates telomerase also has the potential to feed cancer growth – not quite the healthful result that one might desire! On the flip side, telomerase inhibitors may prove to be a useful treatment for cancer. As of the writing of this textbook, at least one telomerase inhibitor is in clinical trials for certain types of cancer.
Media Attributions
- Eukaryotic chromosomes © Online Open Genetics is licensed under a CC BY-NC-SA (Attribution NonCommercial ShareAlike) license
- Telomerase © Wikipedia is licensed under a CC0 (Creative Commons Zero) license

