Protein structure
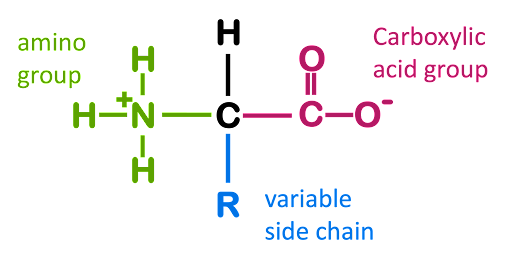
Amino acids are the molecular building blocks used to assemble proteins. The basic structure of an amino acid is shown in Figure 1. Every amino acid consists of a central carbon around which four functional groups are arranged: an amino group (-NH3+), a carboxylic acid (-COO–), a hydrogen (H), and an “R” group that is variable from amino acid to amino acid. Under physiological conditions, the amine, the carboxylic acid, and many of the R groups are charged due to ionization in the aqueous environment of the cell.
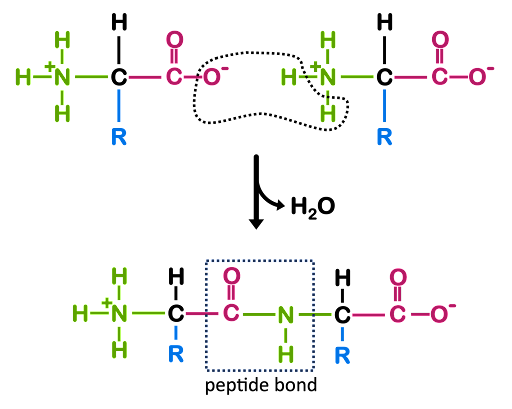
A polypeptide is a polymer of amino acids linked via peptide bond (Figure 2). Figure 2 shows the structure of a dipeptide, but a polypeptide can have hundreds or thousands of amino acids linked together. Once the amino acids are incorporated into a polypeptide chain, they are called amino acid residues.
In Figure 2, you’ll see that the backbone of the dipeptide has the repeating structure -N-C-C-N-C-C-, with each of the amino acid residues contributing one “-N-C-C-“ unit to the repeat. Note that the backbone has polarity: at one end of the structure is a free amino group, and at the other is a carboxylic acid group. All polypeptides, regardless of length, have this same polarity: an amino group at one end and a carboxylic acid at the other. The ends of the polypeptide are therefore called the amino- or N-terminus and carboxyl- or C-terminus. During translation, the ribosome adds subsequent amino acids to the C-terminus of a growing polypeptide chain, so we say that translation proceeds from the N- to C- terminus just like DNA and RNA synthesis proceed from 5’ to 3’.
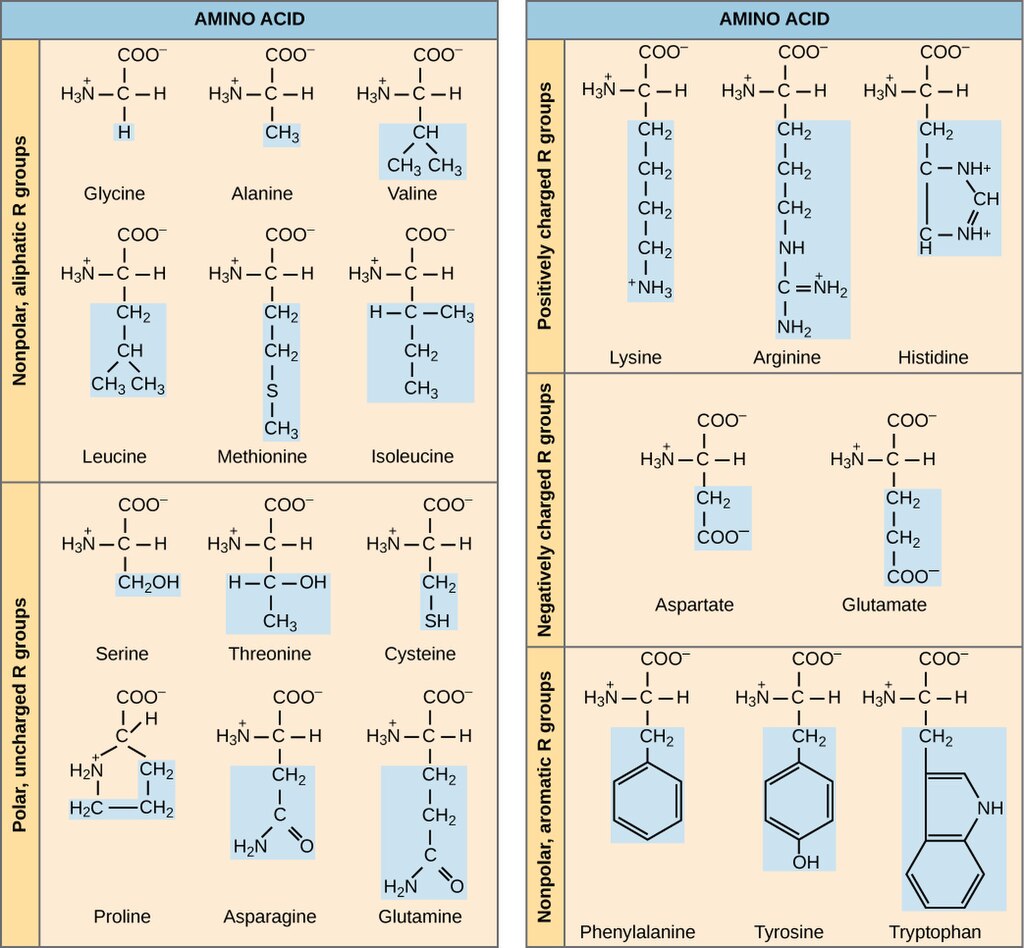
There are twenty amino acids commonly used for proteins in the cell. The names and structures of these are shown in Figure 3. Figure 3 sorts the amino acids based on the chemistry of their side chains. You’ll notice that there are nonpolar, polar uncharged, positively charged, negatively charged, and uncharged aromatic side groups. The chemistry of these side chains affects the structure and function of the protein.
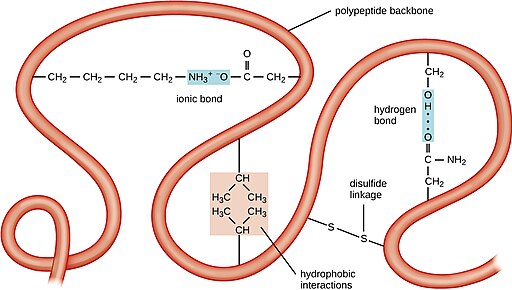
The polypeptide will fold up into a three dimensional structure as it is synthesized, depending on the interactions among the R-groups of the amino acid residues and the backbone of the polypeptide. This is dependent on ionic bonds, hydrogen bonds, covalent bonds, and hydrophobic interactions. Examples of these bonds are shown in Figure 4.
In Figure 4, the backbone of the polypeptide is depicted as a ribbon, with selected amino acid side chains shown participating in folding. You can see an ionic bond between positively charged lysine and negatively charged aspartate, a hydrogen bond between serine and and asparagine, hydrophobic interactions between two valines, and a covalent disulfide linkage between two cysteine side chains. These types of bonds hold the folds of the backbone in place, and the sum of these intramolecular bonds results in a distinctive three dimensional structure for each protein.
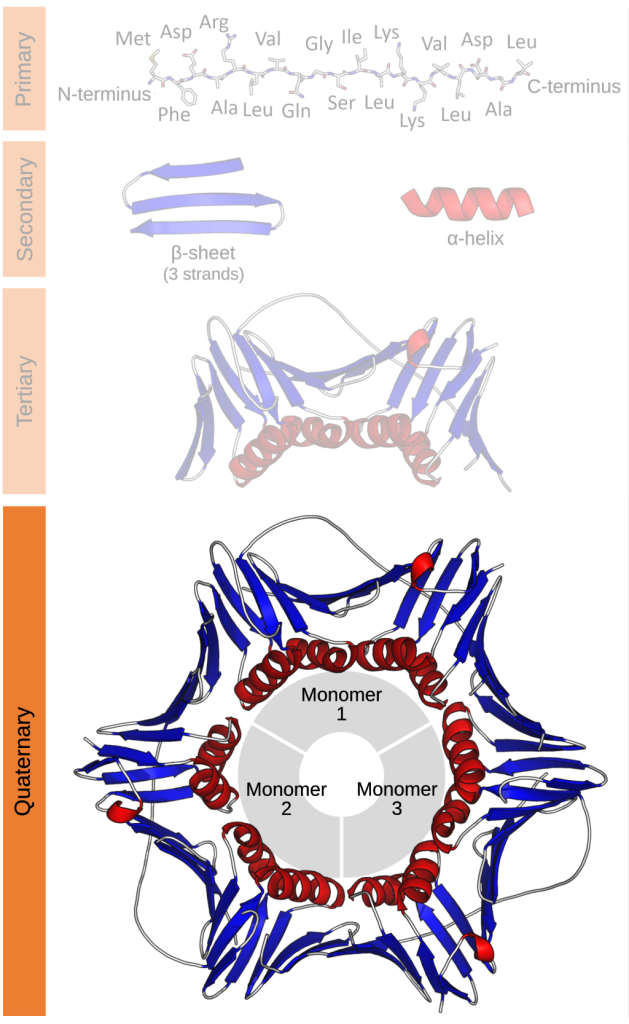
The structure of a protein can be described in terms of its primary, secondary, tertiary, and quaternary structure. The primary (1o) structure is simply the order of the amino acids in the polypeptide. Convention is to list them in order from N-terminus to C-terminus. The secondary structure (2o) refers to the recognizable elements alpha helix and beta sheet within the larger structure. An alpha helix is a region of the protein that folds into a coil, while a beta sheet is a region where the polypeptide backbone folds back and forth in a pleated structure.
Tertiary structure is the full three-dimensional structure of the folded polypeptides. Most proteins have both alpha helices and beta sheets within their tertiary (3o) structure. However, it is not uncommon for a protein to be primarily alpha helices or beta sheets. Some functional proteins are made up of multiple polypeptide chains. These proteins have quaternary (4o) structure. Not all proteins have quaternary structure.
Examples of this hierarchy of protein structure are shown in Figure 5.
Media Attributions
- Amino acid © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license
- Peptide
- Amino Acid structures © Wikipedia is licensed under a CC0 (Creative Commons Zero) license
- OSC Microbio tertiary © Wikipedia is licensed under a CC0 (Creative Commons Zero) license
- Protein structure © Wikipedia is licensed under a CC0 (Creative Commons Zero) license

