Review of transcriptional regulation
In the Overview of Gene Regulation chapter, we saw examples of the levels at which genes can be regulated, from accessing the DNA, transcription of a gene, processing of RNA, and protein synthesis. We ended the chapter with examples of how transcription factors – both activators and repressors – work together to control the expression of prokaryotic genes. As in prokaryotes, eukaryotes use a combination of activators and repressors to control gene expression according to the needs of the cell, although this process is usually much more complicated than in prokaryotes. But there are some other differences as well.
In contrast to prokaryotic genes that might be controlled by one or two regulatory transcription factors, eukaryotic gene regulation is much more complicated. A reminder that the word factor refers to a protein, and the word element refers to a DNA segment.
Remember that prokaryotes can use operons to co-regulate genes in one long RNA transcript with multiple internal ribosomal binding sites and multiple coding sequences. However, this is not possible in eukaryotes where the ribosome binds to the 5’ cap. So instead, eukaryotes co-regulate genes by reusing the binding sites for a specific transcription factor in multiple places in the genome, allowing those genes to be co-regulated. These mechanisms are compared in Figure 3, reprinted from the Overview of Gene Regulation chapter.
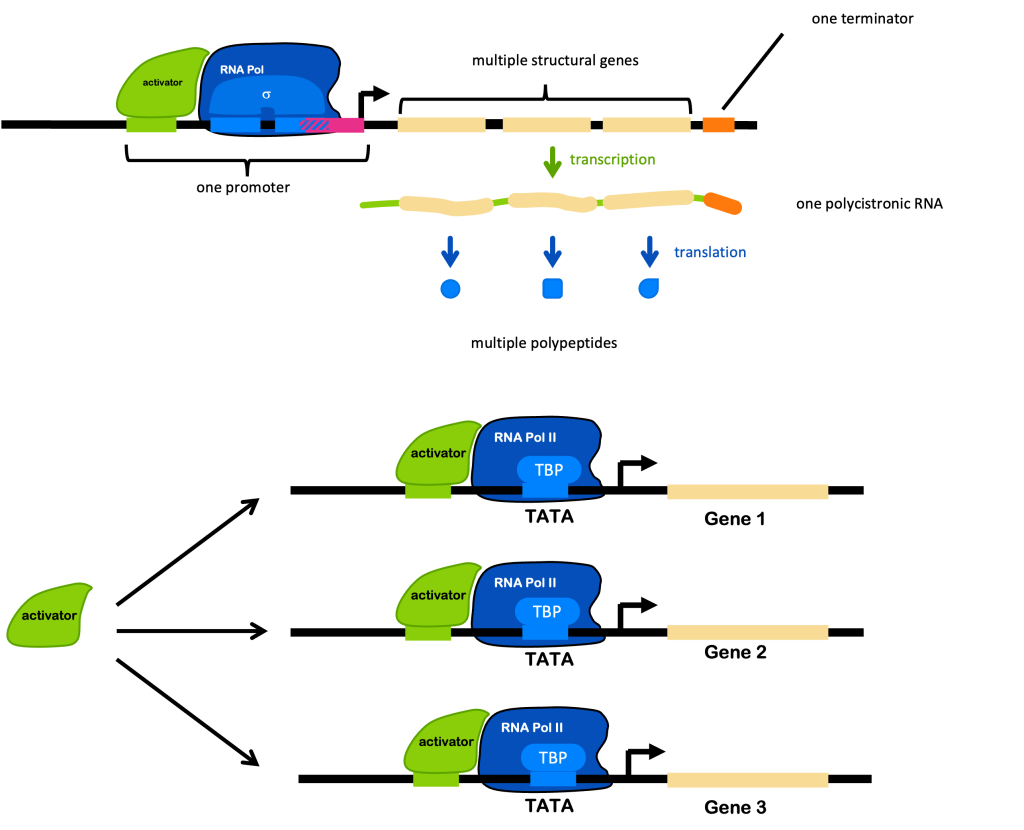
An example: in humans and other mammals, many genes are turned on in response to the hormone estrogen. Those genes have an estrogen response element in the regulatory region of their promoter, which is bound by the estrogen receptors to activate transcription. The estrogen response element would be represented by the green DNA element in the Figure 3 (bottom panel). The green activator in the image represents the estrogen receptor, which acts as a transcription factor.
Enhancer elements regulate transcription in eukaryotes
A eukaryotic gene usually has a core promoter directly upstream of the gene (Figure 4). The core promoter is bound by the general transcription factors, which participate in the transcription of all genes transcribed by a polymerase. As discussed in the Transcription module, these are often named for the RNA polymerase that they assist. For example, the general transcription factors that work with RNA Polymerase II are named TFII_, distinguished from one another by letters.
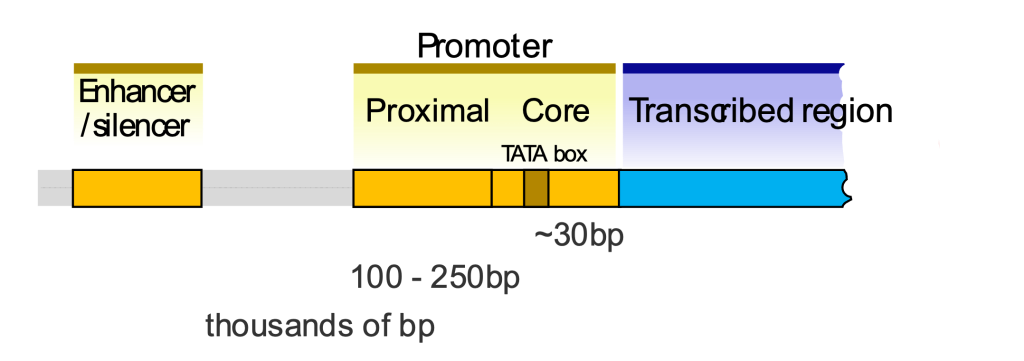
For genes transcribed by RNA Polymerase II, which includes the mRNAs to be translated into protein, the most recognizable feature of the core promoter is the TATA box. The TATA box is bound by transcription factor TFIID in an early stage of transcription initiation. However, additional elements may be present as well, including BRE (TFIIB recognition element), Inr (Initiator), and other elements like the CAAT box and GC box (named for the sequence of bases in the element). Many promoters don’t even have a TATA box!
In prokaryotes, a strong core promoter (-10 and -35 box elements, for example) is sufficient to recruit polymerase and drive expression of an adjacent gene. But in eukaryotes, a core promoter and general transcription factors are not sufficient to drive the expression of eukaryotic genes. Eukaryotic genes require additional regulatory elements. These can include nearby proximal promoter elements, which are bound by specific transcription factors, which are so-named because they are specific to individual genes (or families of genes). They also include distal elements like enhancers and silencers, discussed more in the next section.
These distal elements can be thousands of base pairs from the transcriptional start site, but they may be brought into proximity to the core promoter through the bending of the DNA, as shown in Figure 5, reprinted from the Transcription and Overview of Gene Regulation chapters.
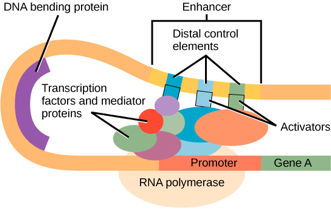
As suggested by the names, enhancers typically function to enhance the transcription of a gene when bound by appropriate factors. At the same time, silencers reduce the transcription of a gene when bound by appropriate factors. A single enhancer or silencer typically has binding sites for multiple factors.
Figure 5 shows three activator-binding elements in one enhancer, but enhancers can include dozens of binding sites for both activators and repressors. The repressors may block the assembly of the polymerase, the mediator, or even other transcription factors. The factors that bind cooperatively to an enhancer are collectively called the enhanceosome.
For some genes, regulatory promoters and enhancers may be pretty straightforward: for example, a gene that is always expressed in one particular cell type (and only that cell type) might have a small number of regulatory elements that control its expression.
But other genes are expressed in complex patterns: for example, they might be needed in multiple tissues (but not all tissues) throughout the adult organism and/or needed at particular times during development. Such genes may have multiple enhancers that control expression. Usually, only one enhancer needs to be active for the gene to be transcribed, but each enhancer might respond to different conditions. In this way, enhancers can act like a network of switches so that a gene might be expressed under condition A, condition B, or condition C.
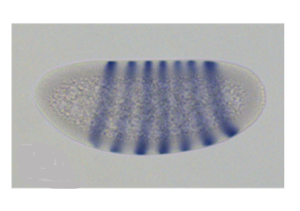
A well-studied example of this is the eve gene from the fruit fly Drosophila melanogaster. The eve gene is expressed in multiple locations along the length of the embryo, as shown in Figures 6 and 7. Figure 6 [1]shows an example of an embryo that has been stained for eve mRNA. The anterior of the embryo is oriented to the left, the posterior is to the right, the dorsal side is on top, and ventral on bottom. Purple marks where eve RNA is produced.

In Figure 7, the genomic region around the eve gene is diagrammed with the transcriptional start site indicated by a right-facing arrow and the enhancers numbered according to the stripe(s) that each controls. The eve gene has five enhancers, each of which “turns on” expression in one or two stripe-like regions of a fruit fly embryo.
The mechanism for their activation is discussed more in detail in later sections. But one interesting thing to note is that the enhancers are found both upstream and downstream of the gene. This is typical of many eukaryotic genes, where enhancers do not need to be directly proximal to the core promoter due to the looping of DNA during transcriptional activation. Sometimes enhancers can be in introns. There are even rare examples of enhancers within exons for other genes!
Later sections of this chapter look at additional examples of how enhancers work in combination to drive complex patterns of gene expression in a multicellular organism. We will look at genes that, like eve, are expressed during embryonic development. We will look first at the expression of genes important for body patterning in Drosophila melanogaster and then at related genes in other organisms (including humans).
Test Your Understanding
Media Attributions
- Figure 3 EG Regulation Operon vs eukaryotic coregulation 2 © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license
- Figure 4 EG Regulation © Thomas Shafee is licensed under a CC BY (Attribution) license
- Figure 5 EG Regulation © CNX OpenStax (Biology) is licensed under a CC BY (Attribution) license
- Figure 6 EG Regulation © Modified from Peterson et al is licensed under a CC BY (Attribution) license
- Figure 7 EG Regulation © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license
- Modified from Peterson et al, https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0004688 ↵

