RNA processing of RNA pol II transcripts
The poly-adenylation of RNA Pol II transcripts is one example of the RNA processing that occurs in eukaryotes. Before processing, the RNA Pol II transcript is called the primary transcript, or pre-mRNA. Only after processing is complete is it described as an mRNA.
Primary transcripts produced by RNA Pol II undergo three main types of processing:
- 5’ capping
- Splicing
- Poly-adenylation (discussed in the previous section)
These modifications of the pre-mRNA are sometimes called post-transcriptional modifications, although this is not a very good description because all three types of modifications are made to RNA as it is still being transcribed. Modification of the transcripts happens in the nucleus, but after processing the mRNA molecules will be exported to the cytoplasm for translation/protein synthesis.
5’ capping
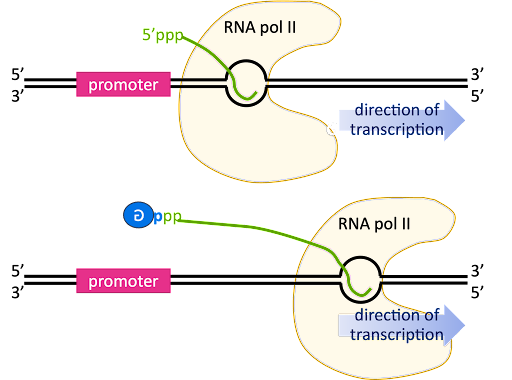
As transcription transitions to the elongation phase, the transcription bubble is translocated down the length of the gene, and the 5’ end of the RNA is displaced from the template strand and extends outward from an exit channel in the RNA polymerase.
The pre-mRNA initially has a triphosphate at the 5’ end, since the beta and gamma phosphates are only lost from a nucleotide when it is added to the 3’ end of a growing chain (review Figure 2 for a reminder!).
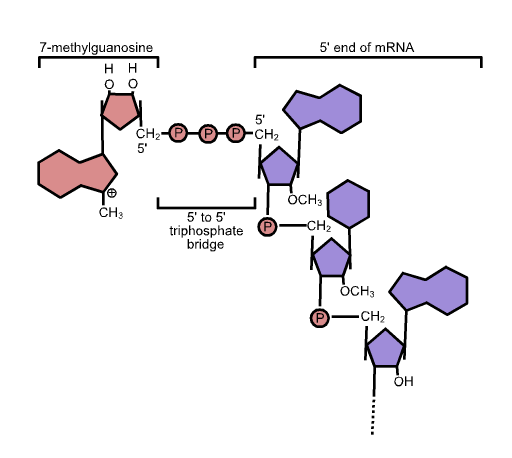
The 5’ end is capped by the addition of a G nucleotide. The capping reaction connects the 5’ end of the RNA to the 5’ position of the capping guanosine, so it is sometimes described as “inverted” or “backward”. In Figure 13 this is indicated by the backward “G” in the blue circle.
The capping reaction retains the alpha and beta phosphates from the 5’ end of the RNA and the alpha phosphate from the capping guanosine, so there is an unusual linkage of 3’HO-sugar-phosphate-phosphate-phosphate-sugar at the capped end. After the guanine is linked to the RNA, it is then methylated at position 7 (a -CH3 group is added). This chemical structure of the cap is shown in Figure 14.
The 5’ cap serves multiple purposes. First, it participates in the translocation of the mRNA out of the nucleus to the cytoplasm for translation. Second, it protects the mRNA from degradation. And third, it serves as a ribosome binding site during translation.
Interestingly, the 5’ cap is exploited by some viruses that infect eukaryotic cells. Viruses like Influenza A have an RNA genome, and specialized viral RNA-dependent RNA polymerases transcribe using the RNA genome as a template. But the products of viral transcription are not capped by the virus. Instead, the virus “snatches” caps from cellular RNA molecules, along with about 10-15 base pairs of the cellular RNA. These stolen caps are used to prime RNA synthesis to make viral RNAs. Because cap-snatching is unique to viruses – this is not a function uninfected cells perform – the cap-snatching mechanism is a drug target for treatment of viral infections. Inhibiting cap-snatching blocks the viral life cycle without harming the host[1][2].
Splicing
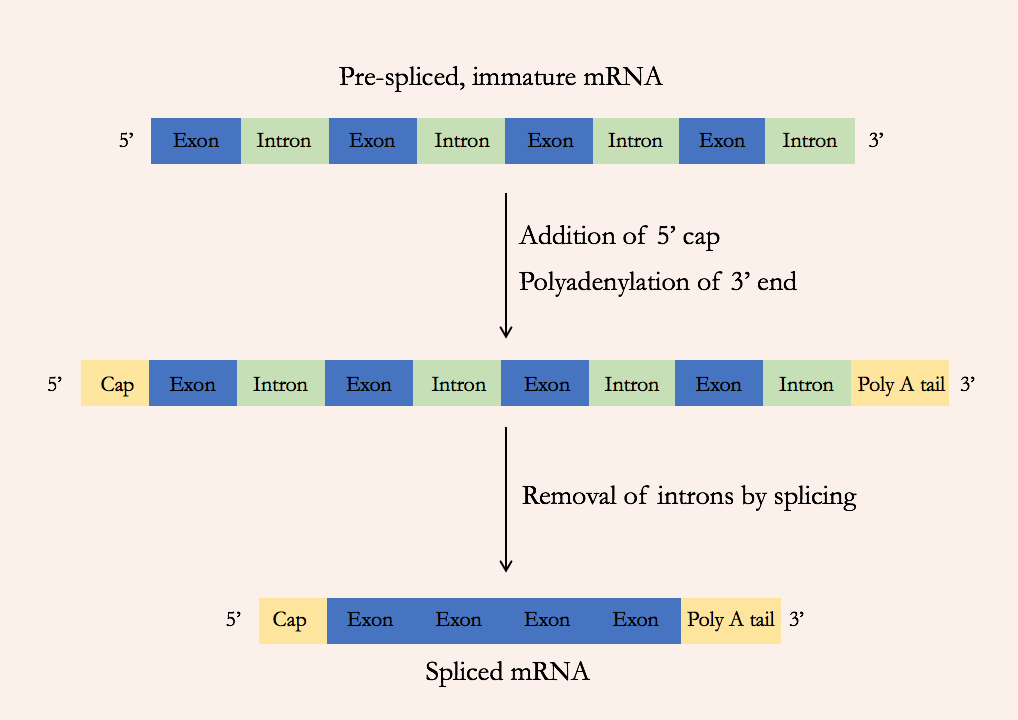
In bacteria, the protein-coding sequence of a gene is present in the genome in one long continuous stretch. But in eukaryotes, the protein-coding sequence can be broken up into many segments within the genome, interspersed with noncoding DNA. The pre-mRNA includes both the protein-coding sections (called exons) and the intervening noncoding sections (called introns). But the mature RNA includes only the exons: the introns are removed during a process called splicing (Figure 15).
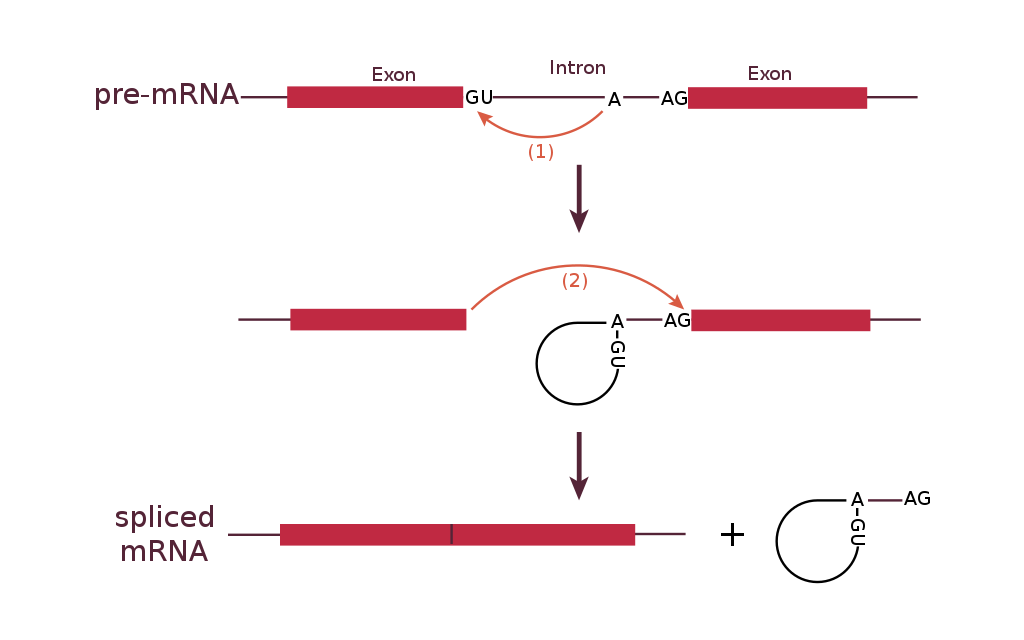
1. A loop is formed by a bond between the branch point A and the 5’ end of the intron. This separates the 5’ end of the intron from exon 1.
2. The 3’ end of exon 1 is connected to the 5’ end of exon 2, releasing the looped intron from the mRNA.
The most common splicing mechanism uses a complex called the spliceosome to remove the introns and connect exons together. The general mechanism of splicing is shown in Figure 16.
Within the intron is a sequence called the branch point. The spliceosome forms a covalent bond between an A nucleotide at the branch point and the first (5’) nucleotide of the intron. This breaks the sugar-phosphate backbone at the 5’ intron/exon boundary and forms a loop, or a lariat, in the intron.
A new phosphodiester bond is then formed between the newly exposed 3’-OH group of the first exon and the 5’ phosphate at the beginning of the second exon, at the 3’ intron/exon boundary. This releases the intron from the second exon as a lariat structure. The lariat is later degraded in the cell.
Like the other processes of molecular genetics, splicing is regulated by the interaction between sequence elements and regulatory factors.
The spliceosome is a large multi-subunit complex assembled from multiple snRNPs, pronounced “snurp”. snRNP stands for “small nuclear ribonuceloproteins.” The spliceosome snRNPs are named U1, U2, U4, U5, and U6. (There is also a U3 in the cell, but it does not participate in splicing).
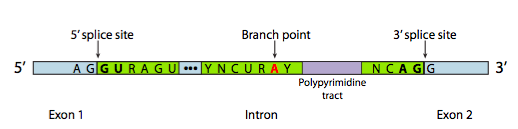
Each snRNP has an RNA and a protein component. The snRNPs bind to consensus sequences at the intro/exon boundaries via complementary base pairing between the snRNAs and the pre-mRNA. These sequences are called the 5’ and 3’ splice site, after their orientation at the 5’ and 3’ ends of the intron. snRNPs also bind to the region of the RNA around the branch point A. The three sequences are shown in Figure 17[3].
The sequence at the 5’ splice site, the branch point, and the 3’ splice site can vary from intron to intron. Even multiple introns from the same gene will not have identical sequences. But some of the bases are highly conserved: nearly every eukaryotic intron begins with the bases “GU” and ends with the bases “AG”. These bases are bolded in Figure 17.
Test Your Understanding
The spliceosome-mediated splicing described here is a common mechanism of splicing. But it is worth mentioning that some introns are spliced via other means. Some introns are spliced via another spliceosome that uses different snRNPs. And some introns are self-splicing! Remember that RNA can act as an enzyme. Self-splicing introns catalyze their own removal from the pre-RNA.
Alternative splicing
Most eukaryotic genes have at least one intron, but some have many more. The human dystrophin gene has 78 introns!
In some cases, a single gene can be used to produce multiple RNAs. This is accomplished through alternative splicing. With alternative splicing, not all exons are incorporated into a mature mRNA. Which exons are included are regulated by a collection of splicing factors, which block the use of some 5’ and 3’ splice sites and promote the use of others. Alternative splicing is illustrated in Figure 18. In Figure 18, a gene with five exons is transcribed into pre-mRNA. The pre-mRNA can be spliced to include all five exons or alternatively spliced to skip one or more exons. The alternatively spliced mRNA will encode different forms of the translated protein.
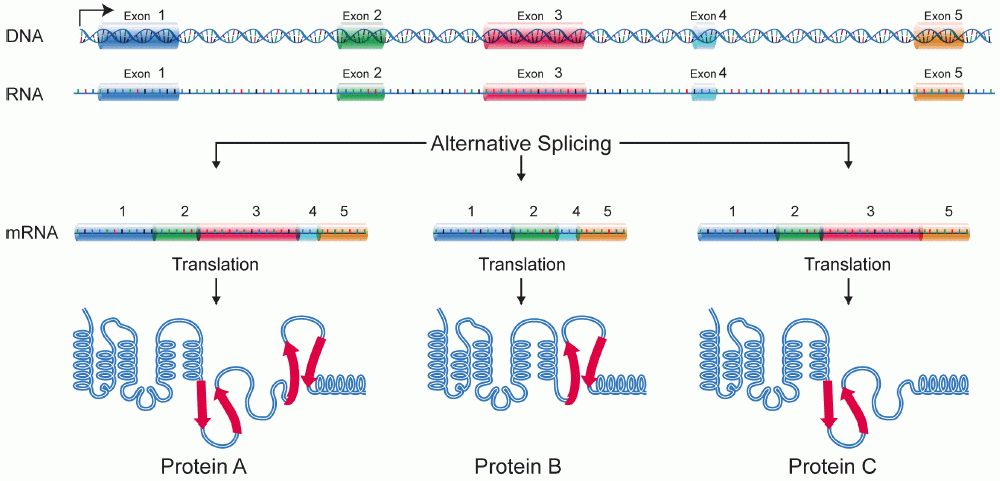
Alternative splicing builds versatility and efficiency into the genome: only one gene is needed to produce multiple proteins. Alternative splicing is also an exception to the “one gene, one polypeptide” rule discussed at the beginning of this module: with alternative splicing, one gene can produce several different polypeptides.
Additional forms of RNA processing
Although not discussed extensively in this text, RNA can undergo additional forms of post-transcriptional modification. RNAs can be edited through the addition or deletion of bases or by converting one base to another. This includes the modification of bases to incorporate non-canonical (unusual) bases. tRNAs tend to have many modified bases, including the non-canonical structures like inosine and pseudouracil. Inosine is formed by the modification of adenosine. Psuedouracil is formed by modification of uracil.
Test Your Understanding
Media Attributions
- Capping © Amanda Simons is licensed under a CC0 (Creative Commons Zero) license
- Structure of the cap © Wikipedia is licensed under a CC0 (Creative Commons Zero) license
- Messenger RNA maturation © Wikipedia is licensed under a CC0 (Creative Commons Zero) license
- RNA splicing reaction © Wikipedia is licensed under a CC0 (Creative Commons Zero) license
- Consensus sequences © LibreTexts is licensed under a CC0 (Creative Commons Zero) license
- DNA alternative splicing © Wikipedia is licensed under a CC0 (Creative Commons Zero) license
- Samji, T. Influenza A: Understanding the Viral Life Cycle. Yale J. Biol. Med. 82, 153–159 (2009). ↵
- Noshi, T. et al. In vitro characterization of baloxavir acid, a first-in-class cap-dependent endonuclease inhibitor of the influenza virus polymerase PA subunit. Antiviral Res. 160, 109–117 (2018). ↵
- Book: Cells - Molecules and Mechanisms (Wong). Biology LibreTexts https://bio.libretexts.org/Bookshelves/Cell_and_Molecular_Biology/Book%3A_Cells_-_Molecules_and_Mechanisms_(Wong) (2018). ↵

