Successive action of maternal effect, gap, pair-rule, segment polarity, and homeotic genes
You may recall that, during fertilization, the sperm mostly just contributes DNA to the zygote, while the egg contributes both DNA and cytoplasmic components.
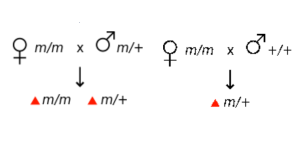
The cytoplasm of the fertilized egg contains maternal mRNA. After fertilization, the maternal mRNA can be used to produce protein quickly before gene expression begins from the zygotic genome. Because the genotype of the mother influences the phenotype of the zygote (not the genotype of the zygote itself!), these are called maternal effect genes. Bicoid is an example of a maternal effect gene. As shown in Table 1, maternal effect genes show a non-Mendelian pattern of inheritance because the phenotype is not dependent on the genotype of the individual.
| All offspring show the wild-type phenotype | All offspring show the mutant phenotype |
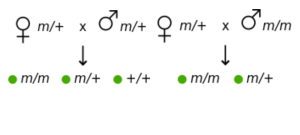 |
 |
Test Your Understanding
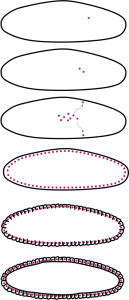
The Drosophila embryo is different from a mammalian embryo in that the earliest cycles of mitosis happen very rapidly – as fast as 8 minutes per cycle – but do not include cytokinesis (the separation of the cytoplasm)(Figure 20). There is also not much growth in size. As a result, the early embryo consists of a syncytium (pronounced “sin-sish-um”) multiple nuclei together in a continuous cytoplasm. Within a few hours of fertilization, the nuclei migrate to the outer edges of the syncytium. The cytoplasm is compartmentalized to form individual cells.
Bicoid mRNA is localized to the anterior end of the syncytium. As the embryo translates the mRNA, Bicoid protein diffuses throughout the syncytium, forming a gradient with the highest concentration at the anterior end of the embryo. Other maternal effect genes have different distribution patterns throughout the embryo. For example, Nanos protein is distributed in a gradient with the highest concentration at the posterior end. And other proteins are important for dorsal-ventral patterning.
The bicoid protein is a morphogen that triggers the development of structures within the embryo through a cascade of gene expression events. Bicoid protein can activate zygotic genes where it contacts the zygotic nuclei within the syncytium. As individual cells pinch off parts of the shared cytoplasm to become separated from one another, they end up with decreasing amounts of Bicoid protein toward the posterior of the embryo. The different concentrations of bicoid in the separated cytoplasms lead to differential expression of bicoid-regulated genes (like eve).
Bicoid directly or indirectly regulates genes involved in the segmentation of the embryo, including hunchback, which is one regulator of the eve gene discussed earlier. Although maternal hunchback RNA is distributed throughout the embryo, it is only translated toward the anterior end of the embryo due to translational repression by the Nanos protein in the posterior. Soon after fertilization, though, the zygotic hunchback gene is transcribed, driven by multiple factors. Bicoid drives the production of Hunchback protein at the anterior of the embryo, while a second region of Hunchback expression occurs at the posterior end of the embryo, driven by bicoid-independent mechanisms.
Hunchback belongs to a class of genes called gap genes because they are expressed in broad swaths of the embryo, and loss-of-function mutations result in embryos with a gap of missing structures. Bicoid and hunchback, in turn, participate in regulating the expression of kruppel, giant, and knirps, which are three other gap genes.
In the previous section, we looked at how Bicoid, Hunchback, Kruppel, and Knirps regulate the expression of even-skipped (eve). Eve belongs to a class of genes called pair-rule genes, along with other genes like odd-skipped, hairy, and fushi-tarazu. Like eve, these genes are expressed in alternating stripes along the length of the embryo. The expression of the pair-rule genes are dependent on maternal effect and gap genes, as well as interactions with each other.
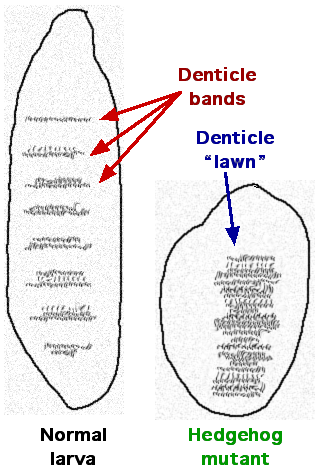
The pair-rule genes, in combination with the maternal effect and gap genes, then influence the expression of segment-polarity genes, which establish a difference between the anterior and posterior edges of segments in the Drosophila embryo. Some examples of segment-polarity genes include the Drosophila genes hedgehog (Hh) and gooseberry (gsb). These genes were also named for the embryonic phenotypes linked with mutations in these genes. A drawing of a hedgehog mutant embryo is shown in Figure 21; rather than the orderly arrangement of denticles seen in a wild-type embryo, hedgehog mutant embryos had a lawn of denticles scattered across the surface of the embryo. Remember that denticles are small projections that extend from the surface of the embryo. Perhaps you can imagine why the mutation was named “hedgehog.”
Finally, the segment polarity genes, pair-rule genes, gap genes, and maternal effect genes collectively regulate the segment identity genes. These genes include the homeotic genes like antennapedia (Antp) and ultrabithorax (Ubx) that were described at the beginning of this chapter. These genes do not affect segment number; they just affect the identity of the segment.
The homeotic genes (or Hox genes) also encode transcription factors. These factors share a common amino acid sequence called a homeobox. The homeobox is a DNA binding domain that allows these factors to recognize elements that regulate the development of body structures. They control the expression of the genes needed to build structures like wings, eyes, and antennae.
For example, the expression of antennapedia is important for the development of leg structures. But if the antennapedia is mis-expressed in the head, leg structures grow in place of the antenna. This is shown way back in in Figure 2, and is what gives the gene the name of antennapedia. Where ultrabithorax is expressed in thorax and abdominal segments, it represses wing formation. A loss of function in ultrabithorax results in the additional set of wings we see in Figure 1, while a gain of function mutation prevents the normal forewing structure from developing. Expression of a gene called eyeless in the leg leads to the formation of eye structures on the leg.
Together, the classes of genes described here exemplify spatiotemporal gene regulation, where the timing of gene expression and the position of a cell within an organism determine which genes are expressed. In early embryonic stages, each class of genes is dependent on the combinatorial effects of earlier-expressed classes. In later stages of development, these gene products together influence patterning in specific organ systems.
Test Your Understanding
Media Attributions
- Table 1-Wild-cropped © Celefin is licensed under a CC BY-SA (Attribution ShareAlike) license
- Table 1-Mutant-cropped © Celefin is licensed under a CC BY-SA (Attribution ShareAlike) license
- Figure 20 EG Regulation © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license
- Figure 21 EG Regulation © JWSchmidt is licensed under a CC BY-SA (Attribution ShareAlike) license

