The histone code
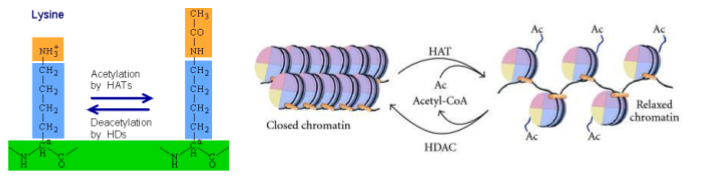
Acetylation of histones is associated with increased gene expression
Acetylation of lysine covalently links an acetyl group (-COCH3) to the amine group (-NH3+) in the lysine sidechain as shown in Figure 5a. This makes the side chain being less positive, which in turn loosens the interaction between DNA and histones and relaxes chromatin structure (Figure 5b).
Acetylation of lysines is catalyzed by a class of enzymes called histone acetyl transferases, or HATs for short. Acetylation is a dynamic process, and acetyl groups can be removed by enzymes called histone deacetylases, or HDACs for short. In general, acetylation is associated with loose chromatin structure in actively transcribed regions of the genome. Parts of the genome that are not actively transcribed are generally associated with lower levels of acetylation. Heterochromatin has little acetylation.
Methylation of histones has varied effects

Histones can also be methylated. Enzymes called histone methyltransferases (often abbreviated as HMTs) catalyze the transfer of one or more -CH3 functional groups to selected lysine or arginine side chains in the histone tails. The structure of mono, di, and trimethylated lysine is shown in Figure 6. The cofactor S-adenosylmethionine (abbreviated SAM or SAMe) serves as the source or donor of the methyl group. Histone demethylases can remove methyl groups.
In contrast to acetylation, which is almost always associated with transcriptional activation, methylation of histones has a more varied effect. For example, methylation of histone H3 at position K4 is generally associated with increased transcription, but methylation of H3 at position K9 is associated with decreased transcription.
Test Your Understanding
Histone modifications are maintained after replication
Histone modifications are maintained in daughter DNA after replication and mitosis. During replication, nucleosomes must be disassembled from the parent DNA to unwind the double helix. Components of the replication machinery help reassemble nucleosomes on daughter strands of DNA, but twice as many nucleosomes are needed post-replication because there is twice as much DNA. Nucleosomes are reassembled on daughter DNA from a mixture of parental histones (which have been modified) and new, largely unmodified histones[1]. The histone code of the parental histones may then be read and re-written to the new histones, although the mechanism by which this occurs is not well understood[2].
Media Attributions
- Histone acetylation © Annabelle L. Rodd, Katherine Ververis, and Tom C. Karagiannis via. Wikipedia is licensed under a CC BY-SA (Attribution ShareAlike) license
- Lysine methylation © Wikipedia is licensed under a CC BY-SA (Attribution ShareAlike) license

