Three-dimensional structures matter
If enhancers are separated from their genes by, in some cases, a million base pairs, and if enhancers may be upstream or downstream of a gene’s transcriptional start site, how do enhancers specifically enhance one gene over another nearby gene?
The rules for this quirk in enhancers are still poorly understood. Still, they are likely strongly influenced by the three-dimensional chromatin structures that assemble in the nucleus of a cell. Although individual chromosomes are not visible under the light microscope except when they are condensed during mitosis, the nucleus of the cell is still highly organized, and the looping of DNA that occurs to regulate transcription is carefully orchestrated. The organization has been described as chromosome “neighborhoods,” with regions of the genome collected in topologically associated domains, or TADs. The word topology refers to shape and structure. For example, a topological map or topo map shows information about the hills and valleys of a region, so this is a fancy way of saying “associated in 3D space”.
Figure 7 shows how those TADs might be organized. Panel A shows a cartoon diagram of how chromosomes tend to stay partitioned in their territory in the nucleus, with loops of DNA in proximity to each other, as shown in Panel B. Those loops allow interactions between genes and their regulatory elements. The dotted lines in Panel C show which DNA elements are brought together through folding of the DNA.
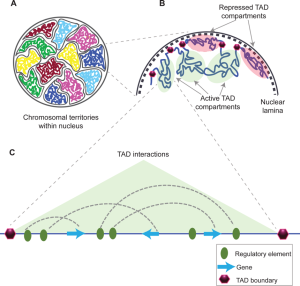
DNA elements called insulators form barriers between genes and nearby unrelated enhancers. A simple model for how insulators function is shown in Figure 8. Looping between the enhancer and the adjacent genes can occur, but the insulators form a boundary beyond which the enhancer cannot act. Insulator sequences are enriched at the boundaries between topologically associated domains and are bound by proteins like CTCF, which helps block the cross-talk of enhanceosomes beyond TAD boundaries.[1]
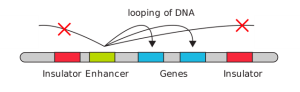
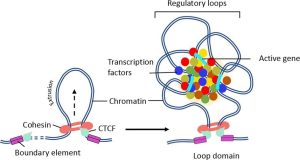
Figure 8. Insulators block enhancers from looping to distal promoters. (Left) Looping between the enhancer and the adjacent genes can occur, but the insulators form a boundary beyond which the enhancer cannot act. (Right) Insulator/boundary elements may be bound by proteins like CTCF. A cohesin ring is wide enough to allow a loop of DNA to extend through, but the loop cannot extend past the CTCF boundary elements. An enhanceosome can assemble with transcription factors binding to loops of DNA brought together in three dimensional space.
Chromatin proteins can interfere with gene expression
In addition, even though we’ve drawn diagrams of genes and enhancers as straight lines and boxes, in the nucleus of a eukaryotic cell DNA is almost always packaged into chromatin and surrounded by proteins. Even during transcription, DNA stays associated with histones, although they must be moved or navigated around for RNA pol to access the DNA.
Just as a repressor protein can block access of an activator to an enhancer, so can chromatin proteins. Two things can’t occupy the same place at the same time, whether an activator is competing with a repressor or whether an activator is competing with a nucleosome.
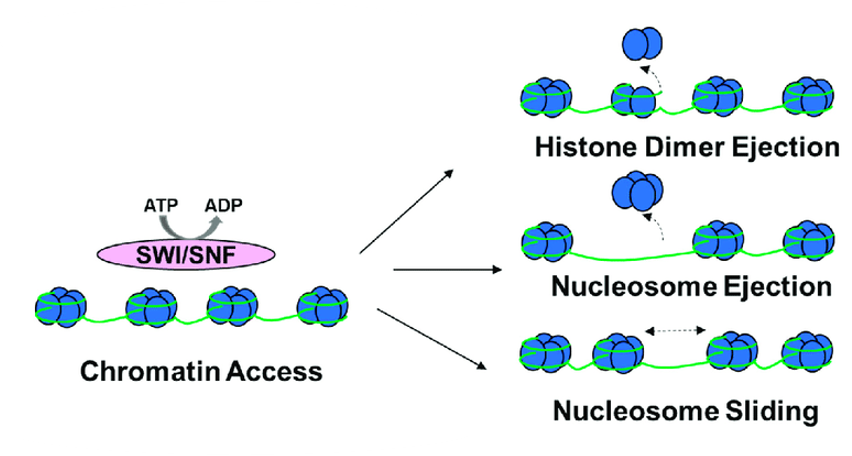
Chromatin remodeling proteins slide nucleosomes out of the way, eject nucleosomes from the DNA, or loosen the association of DNA with histones to allow a segment of DNA to be accessed by the transcription machinery. Some mechanisms of this are shown in Figure 9.[2]
Additional enzymes called chromatin modifiers can covalently modify histone proteins with small functional groups like acetyl or methyl groups. Enzymes called histone acetyltransferases (HATs) and histone deacetylases (HDACs) add and remove acetyl groups, respectively. Enzymes called histone methyltransferases add methyl groups. These modifications make it easier or more difficult to move histones out of the way depending on the modification, shown in Figure 10. [3]
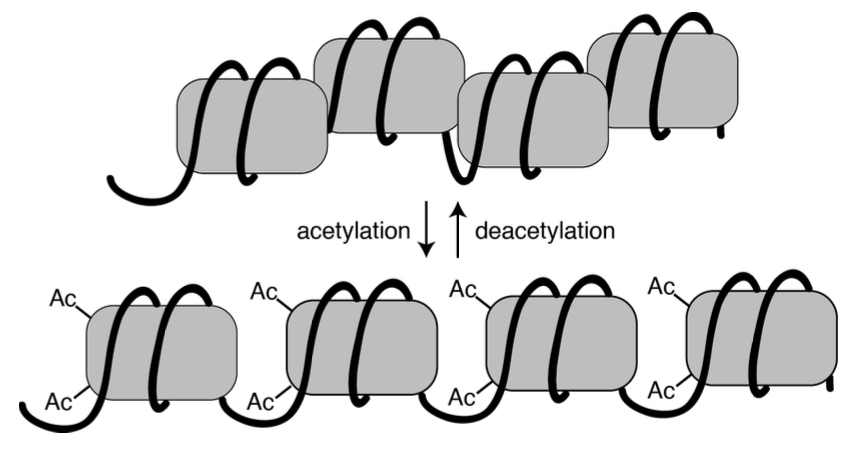
In some cases, these modifications “spread”, with the histone modification state of one part of the genome influencing the modification of nearby nucleosomes. This type of regulation may allow temporal regulation (changes in gene expression over time) of genes that are arranged next to each other along the length of a chromosome.
Some modifications may result in such tight packing of chromatin that transcription cannot occur at all. Tightly packed, transcriptionally silenced chromatin is called heterochromatin, while loosely packed transcriptionally active chromatin is called euchromatin. Certain insulators may serve as barriers to the spread of chromatin modification so that chromatin in adjacent TADs can be in different states.
The influence of chromatin modification in gene expression is discussed in more detail in the chapter on Epigenetics.
Media Attributions
- Figure 7 EG Regulation [TADS] © Navneet Matharu, Nadav Ahituv - Matharu, Navneet (2015-12-03). " Minor Loops in Major Folds: Enhancer–Promoter Looping, Chromatin Restructuring, and Their Association with Transcriptional Regulation and Disease". PLOS Genetics 11 (12): e1005640. DOI:10.1371/journal.pgen.1005640. PMID 26632825. PMC: PMC4669122. ISSN 1553-7404. is licensed under a CC BY-SA (Attribution ShareAlike) license
- Figure 8(Left) EG Regulation © Squidonius (talk) is licensed under a Public Domain license
- Figure 8 (Right) EG Regulation © Pongubala and Murre, Frontiers in Immunology is licensed under a CC BY (Attribution) license
- Figure 9 EG Regulation-Mechanisms of chromatin remodeling. © Modification of Hasan and Ahuja adapted by Amanda Simons is licensed under a CC BY (Attribution) license
- Figure 10 EG Regulation Histones © Deyholos - Open Genetics Lectures adapted by Amanda Simons is licensed under a CC BY-NC (Attribution NonCommercial) license
- Pongubala, J. M. R. & Murre, C. Spatial Organization of Chromatin: Transcriptional Control of Adaptive Immune Cell Development. Front. Immunol. 12, (2021). ↵
- Hasan, N. & Ahuja, N. The Emerging Roles of ATP-Dependent Chromatin Remodeling Complexes in Pancreatic Cancer. Cancers 11, 1859 (2019). ↵
- Locke, J. ‘Open Genetics Lectures’ textbook for an Introduction to Molecular Genetics and Heredity (BIOL207). Borealis https://doi.org/10.7939/DVN/XMUPO6 (2017). ↵

