Tracking linked traits through pedigree analysis
The results of multiple test crosses were historically used to build a chromosome (and genome) map. Hundreds of genes could be mapped in relation to each other in this fashion – even some of Morgan’s initial chromosome maps included nearly one hundred genes.[1]
If a new phenotype was identified, test crosses with known genes could help map the trait to the causative region of a chromosome. This was especially true in organisms where large numbers of offspring could be generated by a single cross. However, test crosses were not useful in organisms where controlled crosses cannot be performed (including humans, where such a thing would be deeply unethical) or for organisms where few progeny might be generated at a time. Test crosses were also less useful for complex multifactorial traits.
In humans, there are many obvious clinical benefits to identifying genes associated with particular phenotypes. By the mid-1950s, geneticists were interested in building linkage groups in humans. Since controlled crosses could not be used, linked traits were tracked through pedigree analysis in large families. They began by looking for phenotypes that could be easily observed and tended to coincide in members of a family.
This sort of pedigree analysis was most useful for traits where both alleles were known for all individuals in a pedigree. So early pedigree analysis often looked for linkage with traits like the ABO blood group: With three different blood type alleles common in human populations, homozygosity and heterozygosity could often be determined from pedigree analysis. One of the earliest linkages found was between Nail Patella Syndrome and the ABO blood group.
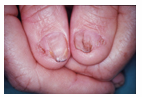
Nail Patella Syndrome is characterized by atypical fingernail growth, missing or misshapen kneecaps, and other developmental differences in the elbow, kidney, and pelvis[2]. An image of the fingernail phenotype is shown in Figure 17. Nail Patella Syndrome is inherited in autosomal dominant fashion. Most examples of Nail Patella Syndrome are familial, but there have been documented examples of NPS patients with de novo mutations that were not inherited from parents.
An example of how NPS and ABO blood type might track in a family is shown in Figure 18.
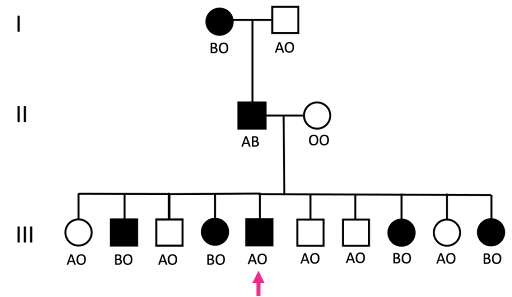
The ABO blood group locus determines whether you have blood type A, B, AB, or O. There are three alleles: A, B, and O, where A and B are codominant, and O is recessive. In the pedigree in Figure 18, Nail Patella Syndrome tracks with the B allele in all individuals except the individual in generation III marked with a pink arrow. The individual marked with a pink arrow has a recombinant phenotype, arising from crossing over between the NPS locus and the ABO locus in the NPS-affected parent in generation II.
This type of pattern is typical of linked traits in a pedigree. With only a small number of people, it is difficult to have much confidence in recombination frequency. But just as Morgan and Sturtevant pooled data from multiple crosses, so can human geneticists pool data from multiple families. When this was done for Nail Patella Syndrome, a recombination frequency of about 10% between the NPS locus and the ABO locus was found, suggesting that the genes are about 10 map units apart. Although not discussed in this chapter, geneticists use statistical tests to determine confidence in their estimates of map distance. If you’re interested in learning more about this, you can find an explanatory video from GnomX on YouTube.
Also like Morgan and Sturtevant, geneticists were able to combine additional data to build a larger linkage map; for example, NPS was later found to also be tightly linked to the adenylate kinase locus[3].
As the tools of molecular genetics were developed, testcross mapping experiments and pedigree analysis might serve as a first step in cloning a gene, meaning to isolate it for further study.
Test Your Understanding
Media Attributions
- Nps © Wikipedia is licensed under a CC0 (Creative Commons Zero) license
- Hypothetical pedigree © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license
- Bridges, C. B. & Morgan, T. H. The Third-Chromosome Group of Mutant Characters of Drosophila Melanogaster. (Carnegie Institution of Washington, 1923). ↵
- Nail-patella syndrome: MedlinePlus Genetics. https://medlineplus.gov/genetics/condition/nail-patella-syndrome/. ↵
- McIntosh, I., Dunston, J. A., Liu, L., Hoover-Fong, J. E. & Sweeney, E. Nail Patella Syndrome Revisited: 50 Years After Linkage. Ann. Hum. Genet. 69, 349–363 (2005). ↵

