Transcription in bacteria
The main enzyme that catalyzes the RNA synthesis reaction is RNA polymerase. In prokaryotes, there is only one RNA polymerase, but in eukaryotes there are multiple RNA polymerases that are each responsible for a subset of RNA types. Transcription depends on the interaction between DNA elements and protein factors involved in transcription.
Initiation
The transcription machinery binds to sequence elements called promoters. In bacteria, the core promoter sequence is usually found about 10 base pairs upstream of the actual transcriptional start site, or toward the 5’ end of the nontemplate strand of the DNA. The bases around a gene are numbered to give geneticists a nomenclature for discussing bases within a gene. The first base that is incorporated into the RNA molecule – the start site of transcription — is called the +1 site. The nontemplate strand of the DNA is treated as sort of a number line, counting negative (-1,-2,-3) upstream from the +1 site into the untranscribed region and counting positive into the transcribed region of the gene (+1+2,+3). This is shown in Figure 6.

Although promoters vary from one gene to another, certain sequence elements are conserved, or similar in many promoters. In prokaryotes, two such DNA elements are the -10 box and the -35 box, so named for their position relative to the +1 transcriptional start site. These are also shown in Figure 6.
The prokaryotic RNA polymerase is made up of five core subunits: two α (“alpha”) subunits, a β (“beta”) subunit, a β’ (“beta prime”) subunit, and an ω (“omega”) subunit. During initiation of transcription, the core polymerase becomes associated with a sixth subunit called σ (“sigma”), forming a holoenzyme.
The role of the σ factor is to recognize promoter sequences, and σ factor disassociates from the polymerase after initiation of transcription. There are several different σ factors, and each of the sigma factors recognize slightly different promoter sequences. One common σ factor in E. coli is called sigma70 (σ70). Sigma70 is sometimes referred to as a “housekeeping” or general purpose sigma factor, but other σ factors have specialized functions like the transcription of stress-response genes, genes involved in nutrient uptake, or genes required for sporulation.
Sigma factors bind to the -10 and -35 boxes. The consensus sequence, or most commonly recognized sequence, for σ70 is 5’TTGACA3’ (-35) and 5’TATAAT’3 (-10), although the actual sequences of individual σ70-regulated genes can vary slightly from the consensus. It’s important to note that the -10 and -35 boxes are bound by sigma as double-stranded DNA, but by convention only the sequence of the nontemplate strand is given when describing elements of a gene.
Test Your Understanding
Elongation
The sequence and orientation of the -10 and -35 boxes specify which strand is to be used as the transcriptional template. RNA polymerase holoenzyme binding leads to the unwinding of the double helix around the promoter. Transcription initiates at the +1 site and synthesis continues with polymerase adding nucleotides to the 3’ end of the growing RNA. We thus say that transcription proceeds in a 5’ to 3’ direction.
Be aware, however, that the polymerase is moving along the template strand toward the 3’ end since the RNA is oriented antiparallel to the template strand. In Figure 5, you can see the 3’ end of the RNA is paired with the template DNA strand. The red arrow indicates the direction of transcription, pointing toward the 3’ end of the RNA and the 5’ end of the template strand.
Unlike replication, transcription does not require synthesis of a primer first. Although DNA polymerases cannot synthesize “de novo”, RNA polymerases can. The RNA polymerase will move toward the 5’ end of the DNA template strand, adding nucleotides to the 3’ end of the RNA molecule as it goes.
Also unlike with replication, the transcription bubble closes from behind and opens ahead as transcription proceeds. This means that the 5’ end of the RNA molecule is displaced from the template strand, extending outward from the transcription bubble. You can see this in Figure 5 as well.
You’ll also note in both Figure 5 and Figure 6 that the RNA molecule is complementary to the template strand – because that is how it is synthesized! The template and the RNA are antiparallel to one another, with the 5’ end of the RNA oriented toward the 3’ end of the template strand. But the sequence of the RNA molecule is identical to the nontemplate strand (just with U’s instead of T’s): same sequence, same 5’ to 3’ orientation.
Test Your Understanding
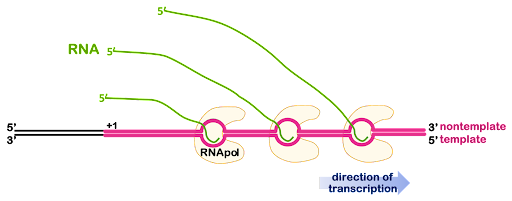
Genes are usually transcribed more than once at a time, producing many RNA molecules. If the RNA will be used for protein coding, having many RNA molecules allows the cell to produce many protein molecules at the same time. Multiple RNA polymerases can act on the same gene, one after another, with additional polymerases beginning transcription before the first polymerase is finished. This is illustrated in Figure 7.
Termination of transcription
RNA polymerase begins synthesis of the RNA at the +1 site and continues until it reaches a terminator element in the DNA. Terminators are the signal sequence in DNA that mark the end of transcription in prokaryotes. There are two types of terminators: Rho-dependent and Rho-independent or intrinsic terminators. Rho-dependent terminators require the action of an RNA helicase called Rho and a sequence in the DNA template that causes the RNA polymerase to pause its elongation of the RNA. Rho unwinds the RNA paired with the template and the RNA is released from the transcription bubble. The polymerase can be recycled and used again to transcribe another RNA molecule.
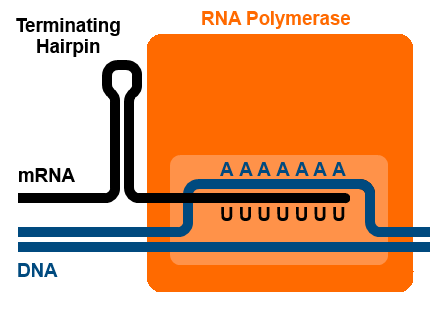
Rho-independent terminators are also called intrinsic terminators. Termination of transcription happens because the RNA structure itself forces the polymerase to release the RNA. RNA molecules with Rho-independent terminators have a GC-rich stretch of base pairs that can fold into a hairpin, followed by a length of U bases. A:U base pairs, like A:T basepairs, are somewhat weaker than G:C base pairs because they form only two hydrogen bonds rather than three. The RNA likely pauses transcription at the A:U base pairs, and the formation of the hairpin destabilizes the RNA:template strand complex enough that the A:U base pairs are not strong enough to hold the RNA and template strands together[1]. This is illustrated in Figure 8.
Media Attributions
- Start site of transcription © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license
- New RNA polymerase © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license
- Terminating Hairpin © Wikipedia is licensed under a CC0 (Creative Commons Zero) license
- Peters, J. M., Vangeloff, A. D. & Landick, R. Bacterial Transcription Terminators: The RNA 3′-End Chronicles. J. Mol. Biol. 412, 793–813 (2011). ↵

