Transcriptional gene expression: Activators and repressors
Whether a gene is transcribed, and how much RNA is produced from a gene, is called transcriptional regulation of a gene. How much RNA is produced is correlated with how readily transcription can be initiated. This, in turn, is correlated with how well the transcription machinery can bind to the promoter of a gene. Cells can “turn on”, “turn up”, “turn down” or “turn off” genes to adjust how much of a particular protein is produced. This allows a cell to react to different environmental stimuli and can allow cells to differentiate, or become specialized.
Recall from the module on transcription: in prokaryotes, transcription is initiated by sigma factor binding to the -10 and -35 boxes, which recruits the RNA polymerase. The “strength” of a promoter is correlated to how well the -10 and -35 boxes match the consensus sequence.
A promoter with a perfect match to the -10 and -35 consensus sequences is called a “strong” promoter. A perfect match to the -10 and -35 consensus sequence (shown in Figure 3) means the promoter can be bound readily by sigma factor. Binding between sigma factor and the -10 and -35 sequences depends on noncovalent bonds between the nucleotide residues and amino acid side chains within the protein. Sigma factor brings RNA polymerase to the promoter, helping it bind to initiate transcription.
A strong promoter is highly transcribed, even without any additional help from other transcription factors. Many RNA molecules are produced from such genes, no additional factors needed (Figure 3, left). For genes with strong promoters, the default expression is “on”. In bacteria, the so-called housekeeping genes, which are always needed regardless of cell conditions, are controlled by strong promoters.
Weak promoters, however, have promoters that are a poor match to the consensus. Changes to the -10 or -35 sequence mean that the sigma factor forms fewer noncovalent bonds with the promoter, and the overall binding between the two is more prone to dissociate, or fall apart, before transcription can begin[1]. Therefore, the gene is transcribed less frequently and the default state is for only low levels of RNA and subsequently protein to be produced (Figure 3, right).
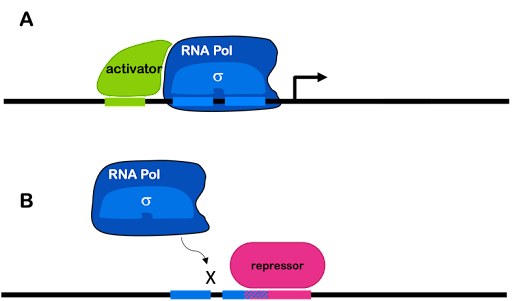
Transcription factors can increase transcription from a weak promoter (in which case they are described as transcriptional activators). When a gene is regulated by an activator,the process is called positive regulation. Also, transcription factors can decrease or block transcription (in which case they are described as repressors). When a gene is regulated by a repressor, the process is called negative regulation. Both strong and weak promoters can be regulated by repressors. A single gene can be regulated by both activators and repressors.
In prokaryotes, activators can stabilize the transcription machinery at a weak promoter by binding both DNA and either polymerase or sigma factor. Repressors commonly bind to DNA and block access of the RNA polymerase holoenzyme to the promoter, preventing transcription. This is often accomplished via the simple mechanism of taking up space! To use an analogy: if my dog is sitting on the couch, there is no room for me. If a repressor is bound to the DNA near the promoter, there is no room for the polymerase.
This is shown in Figure 4. Many activators and repressors bind specific DNA sequences in the promoter of the genes they regulate. Differences in the regulatory regions of promoters plus the activity of different transcription factors are what allow genes to be differently expressed.
Test Your Understanding
Activators and repressors in eukaryotes
Although both prokaryotes and eukaryotes use activators and repressors to control gene expression, the situation is more complex in eukaryotes.
There are no strong eukaryotic promoters: All promoters require additional transcription factors to initiate transcription. In eukaryotes, a large protein complex called the mediator helps assemble the transcription machinery on eukaryotic promoters. Looping of the DNA brings distal elements into proximity with the core promoter. This is shown in Figure 5.
Eukaryotic activators may interact directly with the core polymerase – as is most often true in prokaryotes – but they may also interact with the polymerase indirectly via the mediator or other transcription factors. Activators may bind via proximal elements near to the core promoter, or they may bind to distal elements as part of an enhancer farther away from the promoter.
Each enhancer typically has binding sites for multiple transcription factors, often including both activators and repressors. This allows a combination of factors to coordinate to regulate gene expression under very precise conditions. In Figure 5, you can see that the light-blue element is shared between the enhancers of both genes, but other elements of the enhancer are different. This reuse of sequence elements (and their corresponding protein factor) in multiple genes is common in eukaryotic regulation.
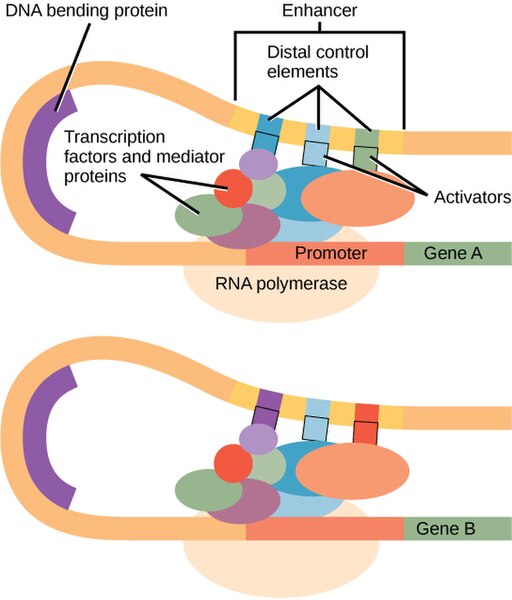
Like their prokaryotic counterparts, eukaryotic repressors may directly block access of the polymerase to the promoter as in eukaryotes. But they may also block an activator from binding to a distal control element in an enhancer, they may interfere with the mediator, or they can alter chromatin compaction in a way that the chromatin itself blocks the transcription machinery’s access to DNA.
Enhancers can be found upstream of a promoter as shown in Figure 5. But some genes are regulated by enhancers found downstream of the coding sequence or even in an intron!
Test Your Understanding
Coordinated expression of multiple genes
In many cases, multiple proteins are needed to work together as a team in a biological pathway. Prokaryotes and eukaryotes use different mechanisms to ensure that all needed proteins are produced at the same time and at appropriate amounts in the cell.
In prokaryotes, functionally related genes are often linked in an operon. These genes are arranged one after the other on the bacterial chromosome, but they are under the control of one single promoter, as shown in Figure 6. The promoter can include binding sites for activators and/or repressors (an activator is shown in Figure 6).
The promoter controls the production of a single mRNA with the coding sequences for multiple structural genes one after another. This is called a polycistronic RNA (a cistron is equivalent to a gene). Each protein is translated independently: each open reading has its own ribosome binding site, start codon, and stop codon. Thus, the genes in an operon are transcribed together, but translated separately. We will see an example of this in the lac operon, described later in this module.
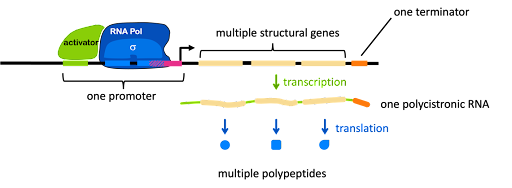
In addition to the structural genes and the promoter, an operon usually has one or more cis-acting elements that are important for gene regulation. Figure 6 shows two such elements: the green element bound by an activator, and a pink element overlapping with the -10 box. Each element is typically recognized by a protein factor that either blocks or facilitates transcription. Note the vocabulary: an element is a segment of DNA, and a factor is a protein.
Test Your Understanding
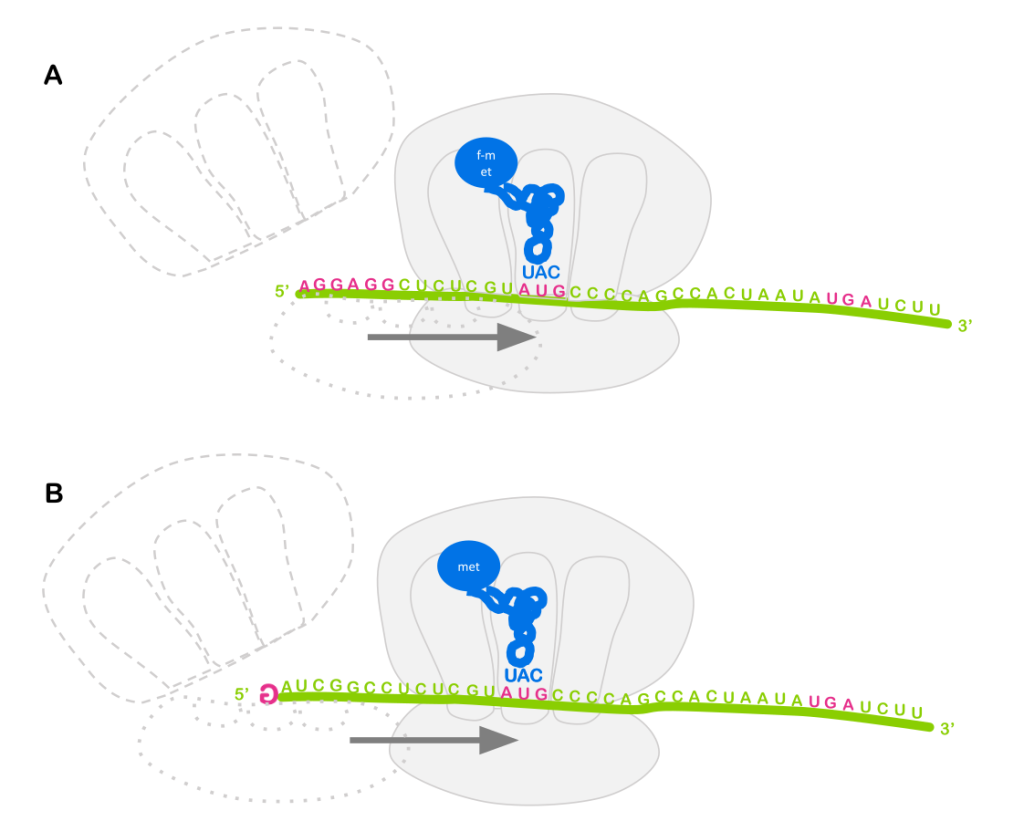
Note: Eukaryotes typically do not use operons to co-regulate genes. You’ll recall from the module on translation that prokaryotes use a ribosome binding site to bring the ribosome to an RNA, and the ribosome binding site positions the ribosome near the AUG start codon for translation initiation (Figure 7A). So it doesn’t matter much whether the ribosome binding site is at the 5’ end of the RNA, or somewhere in the middle: translation will always begin with the nearby AUG, and an RNA molecule can have multiple ribosome binding sites!
But eukaryotes do not generally use ribosome binding sites to attract the ribosome to the RNA. Instead, the ribosome binds to the 5’ cap (Figure 7B). Each RNA only has one 5’ cap, so typically only one open reading frame can be translated per RNA (there are some exceptions to this general rule).
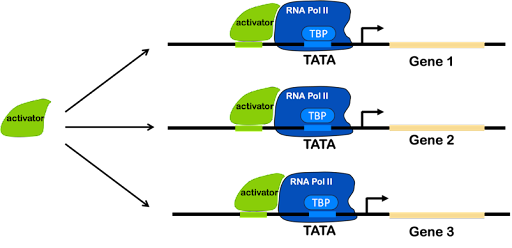
Instead of using operons, eukaryotes use regulatory sequences found in multiple places in the genome. Co-regulated genes have similar elements in their regulatory promoters, which allow them to be activated (or repressed) simultaneously whenever a single transcription factor is present.
Test Your Understanding
Media Attributions
- strong promoter weak promoter
- Regulation of transcription © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license
- DNA bending protein © CNX OpenStax is licensed under a CC BY-NC-SA (Attribution NonCommercial ShareAlike) license
- Operon structure © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license
- Translation initiation © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license
- One activator © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license
- Davis, M. C., Kesthely, C. A., Franklin, E. A. & MacLellan, S. R. The essential activities of the bacterial sigma factor. Can. J. Microbiol. 63, 89–99 (2017). ↵

