Translation in prokaryotes
Like transcription, translation has three stages: initiation, elongation, and termination. In the initiation step, the ribosome must be recruited to the mRNA and positioned over the correct part of the RNA to begin translation in the correct reading frame. The 5’ end of the RNA is not typically translated. During elongation, the ribosome slides along the mRNA from 5’ to 3’, synthesizing a polypeptide to match the codons. During termination, the growing polypeptide is released from the ribosome, and both the ribosome and the mRNA can be reused in another round of translation.
Initiation
In prokaryotes, initiation of translation can begin even before transcription has been completed: Once the 5’ end of the RNA is free of the RNA polymerase, the ribosome can contact the RNA and begin translation as shown in Figure 12. An image of this process as visualized via electron microscope of this process can be seen at Scitable.com.
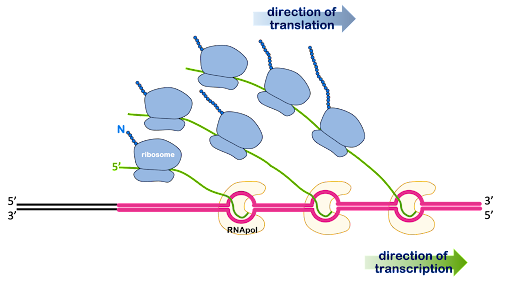
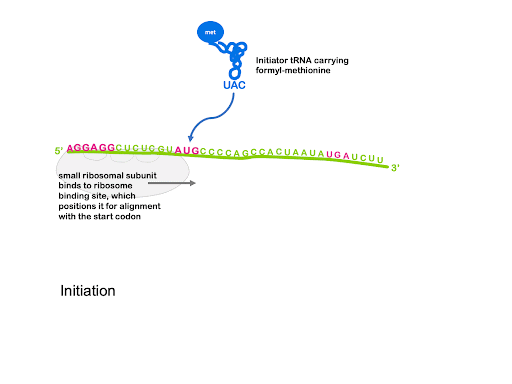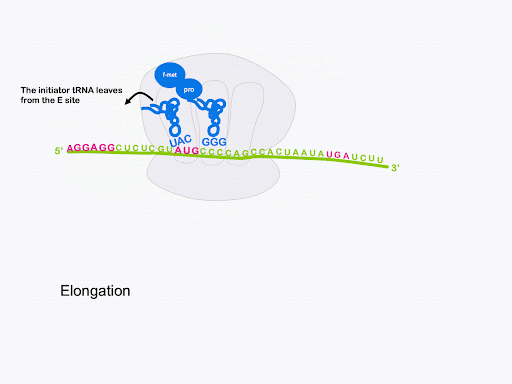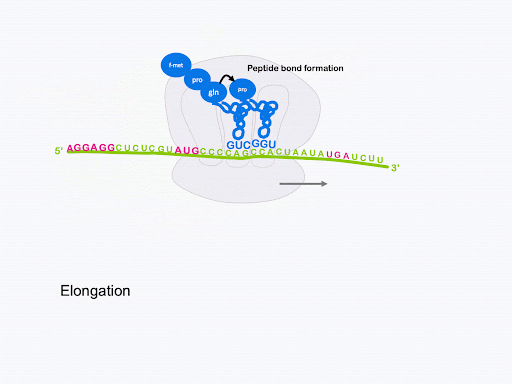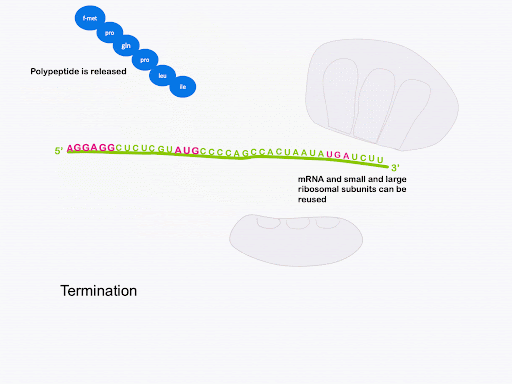
Near the 5’ end of prokaryotic mRNA molecules is a sequence called the Shine-Dalgarno site, or ribosome binding site. This has the consensus sequence of 5’AGGAGG3’. Complementary base pairing between the Shine-Dalgarno site and the sequence 3’UCCUCC5’ within the 16S rRNA of the small subunit is what brings together the mRNA and the ribosome. The Shine-Dalgarno site is positioned to bring the small subunit into proper alignment with the AUG start codon.
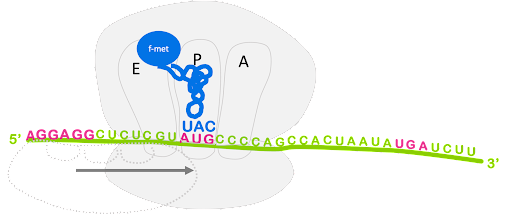
An initiator tRNA charged with f-Met binds to the start codon, and last the large subunit joins, sandwiching the mRNA and fMet-tRNA between the large and small subunit with the initiator tRNA positioned in the P site of the ribosome. This is shown in Figure 13.
Although not shown in Figure 13, the process of translation initiation requires translation initiation factors (IFs) that facilitate binding of the small subunit to the ribosome binding site, prevent premature association of large and small subunit, and help position fMet-tRNAfMet. Translation initiation uses energy from the hydrolysis of GTP to power these steps[1].
Elongation
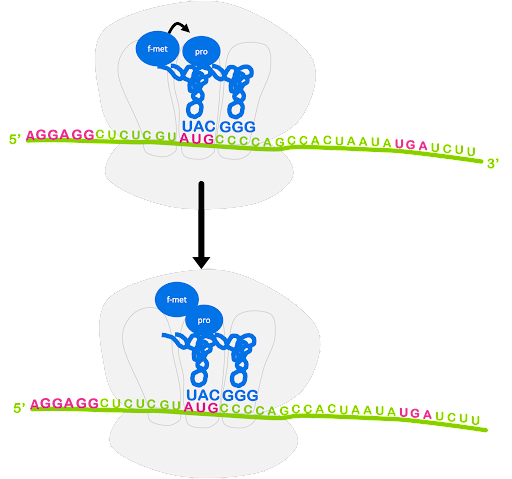
After the large subunit binds, the elongation stage begins. A charged tRNA with anticodon complementary to the next codon will enter the A site of the ribosome, escorted by elongation factor EF-Tu. Again, GTP hydrolysis provides the energy for this process. The ribosome catalyzes the formation of a peptide bond between the first fMet and the second amino acid. In this process, the bond between fMet and tRNA is broken, and fMet is linked with the second amino acid. The second tRNA becomes a peptidyl-tRNA, as it is now linked with a dipeptide instead of a single amino acid. This is shown in Figure 14.
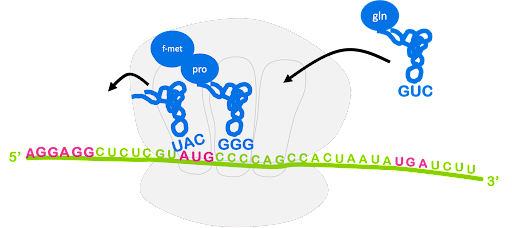
The ribosome translocates, or moves, along the mRNA so that the start codon is positioned in the E site and the original initiator tRNA can exit. The peptidyl-tRNA is in the P site, and the A site is open and can accept a new tRNA. This is shown in Figure 15. Although not shown, GTP hydrolysis by elongation factor EF-G is required for translocation[2].
Termination
The process of bringing in a new charged tRNA, catalyzing peptide bond formation, translocation, and release of the spent tRNA repeats until the entire coding sequence has been translated and the ribosome encounters a stop codon in the A site. There are no tRNAs that recognize the stop codons.
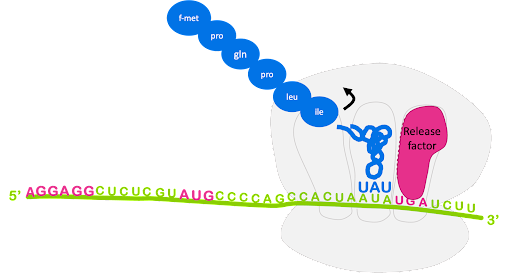
Instead, release factors (RFs) recognize the stop codons in the A site. Release factor binding to the A site promotes the release of the polypeptide from the peptidyl tRNA in the P site. Ribosome recycling factor (RRF) prompts the separation of mRNA, small subunit, and large subunit and recycling of the ribosome (Figure 16). Like initiation and elongation, termination is coupled to GTP hydrolysis[3].
The whole process is shown in the animated gif in Figure 17.
Media Attributions
- Prokaryotes © Amanda Simons is licensed under a CC0 (Creative Commons Zero) license
- Small ribosomal subunit © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license
- Peptidyl transferase reaction © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license
- Translocation of the ribosome © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license
- Termination of translation © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license
- Transcription animation

