tRNAs act as an adaptor between mRNA and amino acid
The ribosome assembles a polypeptide to match the mRNA according to the genetic code. To “read” the mRNA requires the use of a tRNA. tRNAs act as an adaptor between the mRNA and an amino acid. tRNAs form complementary base pairs with the codons of an mRNA, and they carry with them a cognate amino acid.
All tRNAs fold into a characteristic structure. In two dimensions, they are often depicted as a cloverleaf structure, because the backbone of the RNA molecule folds into three stem loop structures as shown on the left in Figure 10. However, their three-dimensional structure is more L-shaped as shown on the right in Figure 10.
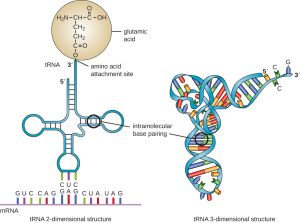
At the bottom of the structure shown in Figure 10 are unpaired bases that make up the anticodon. Each tRNA has an anticodon that can form complementary base pairs with one of the 61 amino acid-specifying codons of the genetic code. In this figure, the tRNA anticodon is CUC, base pairing with the codon GAG, which codes for glutamic acid.
At the top of the structure is the acceptor stem: the region of the tRNA that links to its cognate amino acid. On the left in Figure 10, you can see the tRNA is depicted as “charged” with glutamic acid. The carboxylic acid functional group of the amino acid is covalently linked with either the last 2’OH or 3’ OH group of the tRNA. Once a tRNA has been charged with its matching amino acid, it is more properly called an aminoacyl-tRNA. Enzymes called aminoacyl tRNA synthases perform the charging reaction. Each aminoacyl tRNA synthase is specific for both the tRNA and the amino acid.
Nomenclature: Each amino acid has one or more designated tRNAs. The tRNAs are named for their amino acid, so the one shown in Figure 10 is called tRNAglu. Once it is charged with an amino acid, the amino acid linkage is indicated as well: glu-tRNAglu is glutamic acid tRNA that is carrying a glutamic acid. (Why the double listing of the glu? Under some conditions, tRNAs can be charged with other amino acids.)
The codon-anticodon pairing is antiparallel, just like all base-pairings. And, remember: the codons of a codon table are all written from 5’ to 3’, by convention. So, for example, a UGC codon (Cys) would be recognized by anti-codon 3’ACG5’.
tRNAs have some unusual properties. First, they include non-canonical (unusual) nucleotide residues like inosine and pseudouridine. Remember that bases and nucleoside/nucleotides are named differently; for example, the base adenine is a component of the nucleotide adenosine. Pseudouracil is the base in the nucleotide pseudouridine, and hypoxanthine is the base in the nucleotide inosine. Inosine/hypoxanthine is abbreviated I, and pseudouracil is abbreviated with the symbol psi: Ψ.
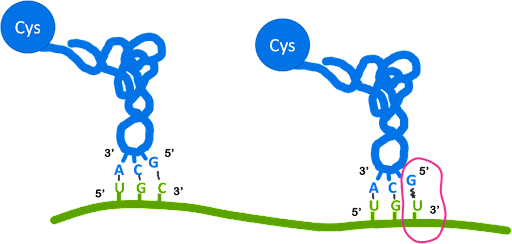
Second, the codon-anticodon pairings do not have to be a perfect match to one another at the third position of the codon-anticodon pairing (3’ codon/5’ anticodon). At this position, noncanonical pairings called wobble base pairs may be used. This allows a single tRNA to recognize multiple codons. The example illustrated in Figure 11 shows that a tRNAcys with anticodon 3’ACG5’ can recognize the codon 5’UGC3’ via canonical base pairing. It can also recognize the codon 5’UGU3’ via wobble base pairing.
Only the 3’ end of the codon, called the wobble position, can utilize wobble pairing. Table 2 lists the base pairings observed at the wobble position. The noncanonical nucleotide inosine is used at the wobble position of some tRNAs, which allows the tRNA to pair with three different codons.
If you look carefully at the codon table shown in Figure 8, you’ll see that the degeneracy of the code is not random. When two codons specify the same amino acid, the codons usually share the same base at the first two positions with either A/G or C/U at the third position. The codons “XXG” and “XXA” can both be recognized by a “XXU” anticodon. And codons “XXC” and “XXU” can both be recognized by a “XXG” anticodon.
| tRNA
5’ end of the anticodon |
mRNA
3’ end of anticodon |
| A | U |
| C | G |
| G | C or U |
| U | A or G |
| I | U, C, or A |
Test Your Understanding
Media Attributions
- tRNA structure © Wikipedia is licensed under a CC0 (Creative Commons Zero) license
- Wobble pairings © Amanda Simons is licensed under a CC BY-SA (Attribution ShareAlike) license

