Overview of Central Dogma and Replication
Objectives
- Describe the chemistry of DNA replication and the function of the players involved, including leading strand, lagging strand, Okazaki fragment, DNA pol I, DNA pol III, primase, primer, sliding clamp, clamp loader, single strand binding protein, helicase, ligase
- Draw a replication bubble and/or replication fork, with leading and lagging strands and all 5′ and 3′ ends labeled.
- Explain why eukaryotic chromosomes get shorter with every round of replication
- Explain the process by which telomerase extends the ends of chromosomes after replication, and recognize that telomerase is only active in certain cell types
Source material
- Replication and Transcription introduction remixed from “Book: Cells – Molecules and Mechanisms (Wong)” by E. V. Wong, LibreTexts is licensed under CC BY-NC-SA. Available free here: https://bio.libretexts.org/@go/page/16085. Selections from Chapter 7 (DNA) and Chapter 8 (Transcription)[1].
- DNA is semi-conservative, remixed from Online Open Genetics (Nickle and Barrette-Ng), available through Biology LibreTexts[2].
Introduction
DNA is a genetic information storage system: it holds the information needed to build an organism. The genetic information storage system has two main requirements. First, the information must be heritable, able to be passed from one generation to the next. Second, the information must be accessible and able to be used by the organism without damaging or destroying the information itself. In the next two modules, Replication and Transcription, we will talk about how these two things are accomplished.
But DNA itself is not a molecule that “does things”. Instead, a collection of other biologically active molecules act upon DNA to read the information and build protein molecules according to instructions in DNA. It is largely the protein molecules that “do” the jobs needed by cells and organisms. So how do we get from DNA to protein? And how is the information shared from parent to offspring?
Recall from the DNA structure module that DNA is composed of two intertwined, complementary strands. If those strands are separated, it’s easy to determine from the sequence of one strand what the sequence of the missing strand should be. You can practice this below.
Test Your Understanding
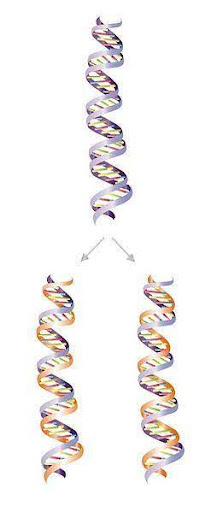
Cells do something similar when they replicate, or copy, their DNA. The strands of the double-helix are separated, and two new strands are constructed by creating a complement for the old ones. The old strands are thus used as templates for new strands (Figure 1). The new strands are called daughter strands. Because each daughter strand retains (or conserves) one intact strand from the parent, DNA replication is said to be semi-conservative.
Thus, two daughter double helices result from one round of replication. When a cell divides, one daughter double helix is passed to each of the two daughter cells. In this way, genetic information is passed from parent to offspring.
So how is the information used? The DNA sequence is not random: it is organized into discrete units called genes. To use an analogy, the characters you are reading on this page are not a random collection of letters, spaces, numbers, and symbols: within the long string of characters are recognizable words and sentences.
Likewise, the genes within DNA contain recognizable sequences that flag parts of the DNA to be transcribed into RNA. During the process of transcription, segments of DNA are read by transcription enzymes, and a molecule of RNA is constructed to be complementary to the parent DNA. In this way, the genetic information stored in the DNA molecule is transferred to a new chemical form. It’s almost like making a photocopy of a page of a library book: the book itself is not altered, but an extra copy of the information is created. The new copy is not exactly the same form as the original – it’s not bound in a hardcover book, it’s only one page and not the whole text, and the paper probably feels a little different – but in many ways it’s also much more useable. You can fold up the photocopy and carry it with you out of the library, you can make notes on it, etc.
Some RNA molecules have specific roles in the cell. Some are enzymes, some regulate cell functions. We will discuss some of these functional RNAs in upcoming modules. But other RNA molecules are specifically used as templates to build proteins. The process of protein synthesis is called translation. To continue with the analogy: if the book you’d photocopied was a cookbook, the protein might be the food you cooked from the recipe.
These three processes together: replication, transcription, and translation, are called the Central Dogma of molecular genetics. The Central Dogma describes the flow of information from DNA: from DNA to daughter DNA, and from DNA to RNA to protein. It is called the central dogma because the ideas provide a framework for nearly all our understanding of genetics.
Since the Central Dogma was proposed and named by Francis Crick in 1957[3], there have been numerous examples of “exceptions to the rule”: other ways information flows through a biological system. Some viruses, for example, including HIV and SARS-Cov2, have an enzyme called reverse transcriptase that creates a DNA molecule from an RNA template. Likewise, RNA molecules can, under certain circumstances, be created using an RNA template. And diseases called prion diseases result from an information transfer from protein to protein. But these exceptions mostly expand our understanding of molecular genetics; they do not counteract our understanding of the flow of information from DNA to protein.
The Central Dogma is illustrated in Figure 2, with the orange arrows indicating the flow of information through the Dogma’s core processes of DNA replication, transcription, and translation. The green arrows indicate additional ways in which information can be transferred. In pink font are listed the molecular machines that perform each process in the cell.
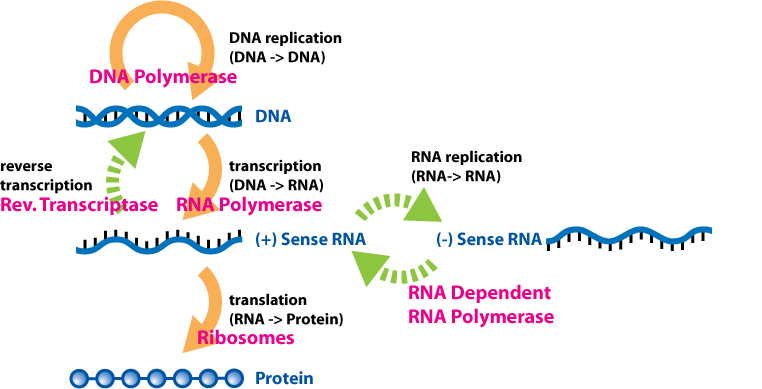
The remainder of this module focuses on the processes of replication and transcription.
- Book: Cells - Molecules and Mechanisms (Wong). Biology LibreTexts https://bio.libretexts.org/Bookshelves/Cell_and_Molecular_Biology/Book%3A_Cells_-_Molecules_and_Mechanisms_(Wong) (2018). ↵
- Nickle and Barrette-Ng. Open Online Genetics. in Open Online Genetics (2016). ↵
- Cobb, M. 60 years ago, Francis Crick changed the logic of biology. PLoS Biol. 15, e2003243 (2017). ↵

